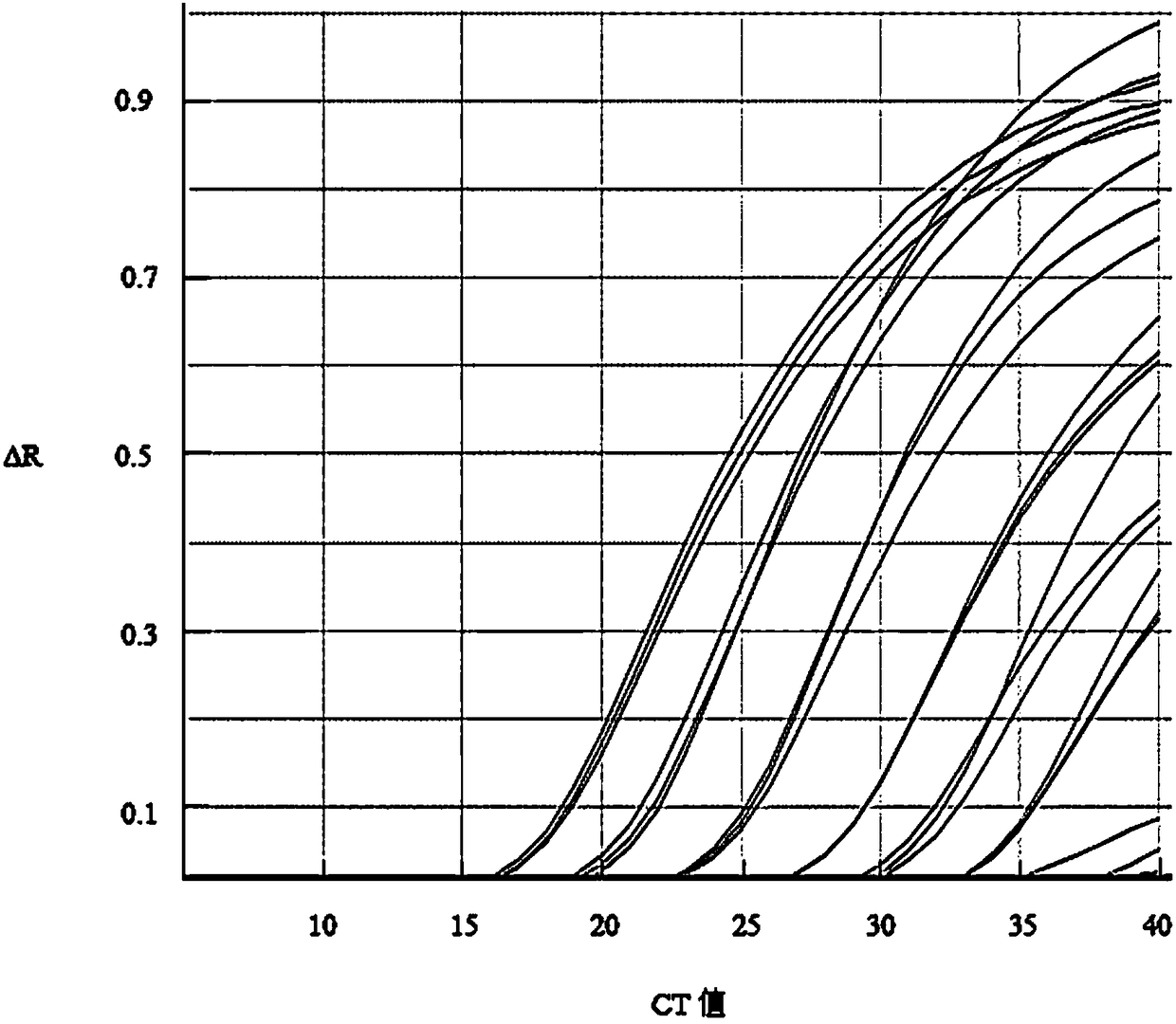
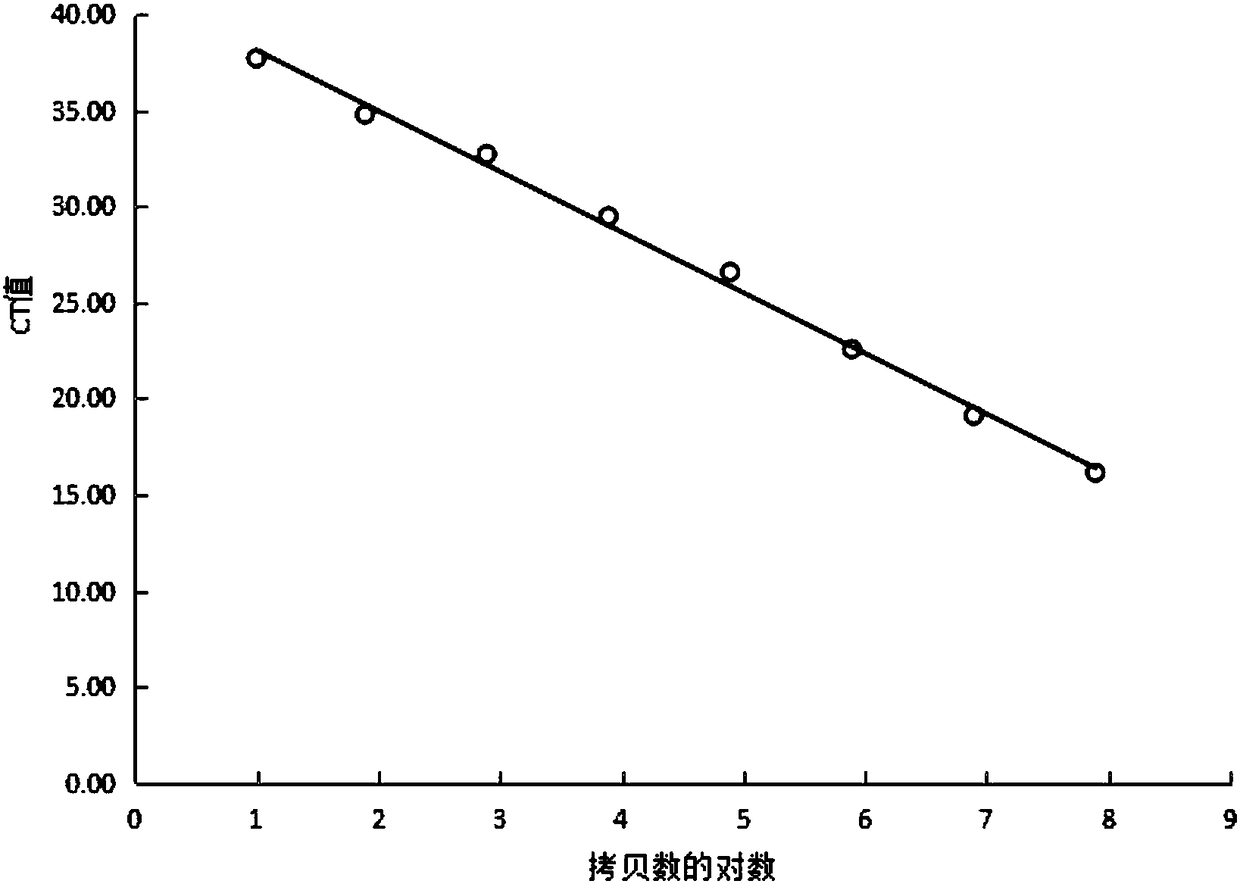
Method for quantitatively detecting white spot syndrome virus through TaqMan probe
A technique for quantitative detection of vitiligo syndrome, applied in the field of molecular biology, can solve the problems of time-consuming results, discrepancies, etc., and achieve the effects of short time, high detection sensitivity, and good application prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] A method for quantitatively detecting prawn white spot syndrome virus using TaqMan probes, comprising the steps of:
[0022] (1) Collect 50 fresh muscle samples of shrimp larvae, put them into alcohol immediately after collection and store them at 4°C.
[0023] Viral nucleic acid extraction: The DNA extraction kit of Tiangen Company was used, and the steps were carried out by referring to the instructions in the kit.
[0024] (2) Design primers and TaqMan probes
[0025] The design of the primer probe was searched for the related sequence of White Spot Syndrome Virus (WSSV) on NCBI, compared with BioEdit software, analyzed the conserved region and specific region of the sequence, determined the primer and probe, and then used Primer Premier5 for primer detection. Needle assessment, initial screening. After synthesizing primers and probes, use PCR to verify whether the target fragment is amplified and the specificity of amplification, and further determine feasible pri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com