Method for quickly and efficiently screening SgRNA (short guide RNA) targeted DNA sequences
A DNA sequence and screening method technology, applied in the field of cell biology, can solve the problems of limited number of detection sites, long time period, cumbersome operation, etc., and achieve the effect of improving efficiency, high success rate, and strong operability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 A method for rapid and efficient screening of sgRNA targeting DNA sequences
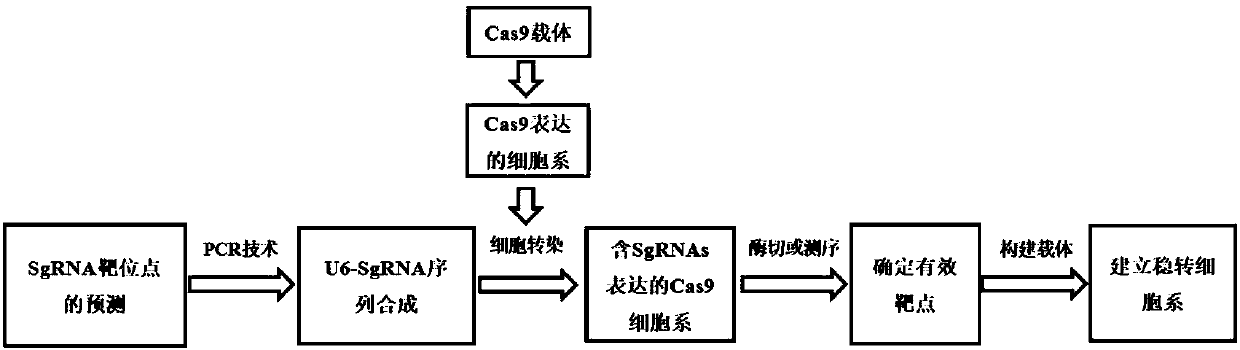
[0032] see figure 1 , is a technical roadmap of a method for quickly and efficiently screening sgRNA targeting DNA sequences of the present invention, comprising the following steps:
[0033] 1. Obtain general template DNA-1 and 2 by PCR method
[0034] The pLVX-hU6-SgRNA vector (purchased from Addgene) was used as a template, and universal PCR amplification was performed with universal primers 1F and 1R, and universal primers 2F and 2R designed for the test detection.
[0035] The amplification primers are:
[0036] 1F: CTTTGGCGCCGGCTCGAGTGTACA (SEQ ID No: 1);
[0037] 1R: CGGTGTTTCGTCCTTTCC (SEQ ID No: 2);
[0038] 2F: GTTTTAGAGCTAGAAATAGCA (SEQ ID No: 3);
[0039] 2R: CACCGGTTAGCGCTAGCTAATGCC (SEQ ID No: 4)
[0040]The general PCR program is: pre-denaturation at 95°C for 3min; 35 cycles of 95°C for 30s, 60°C for 30s, and 72°C for 40s; final total extension at 72°C for 7min; ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com