Molecular marker assisted breeding primer based on growth traits of strongylocentrotus intermedius and screening method
A molecular marker-assisted and growth trait technology, applied in the field of molecular biology, can solve the problem of fewer detection sites and achieve high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
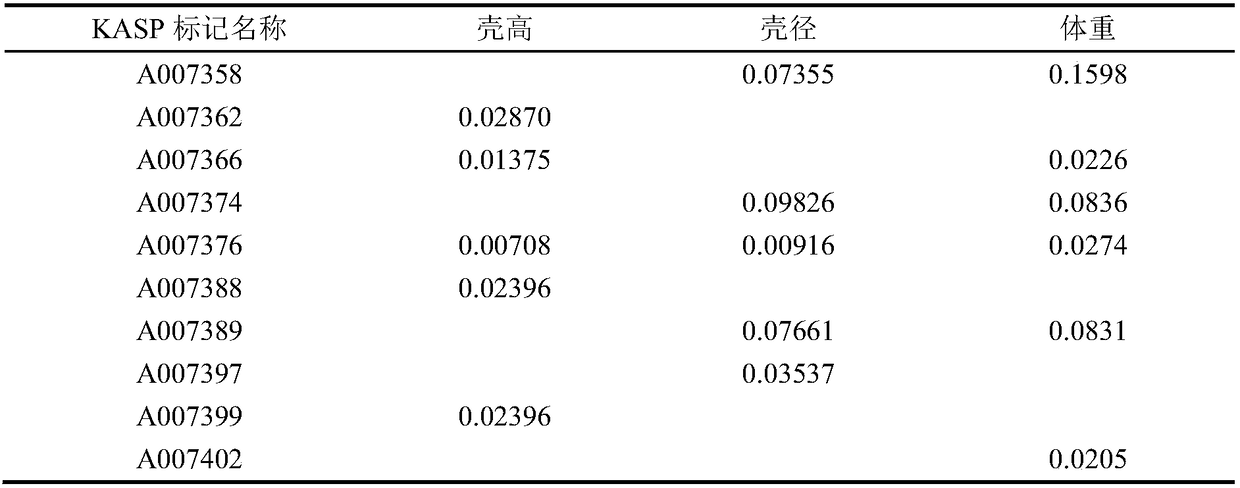
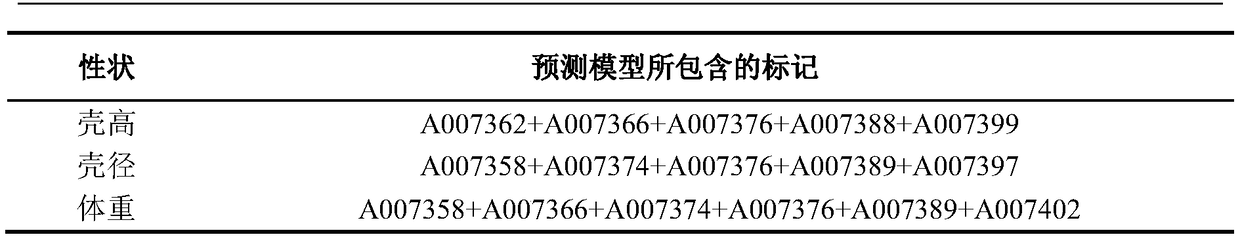
[0034] 1. SLAF tag screening for growth traits
[0035]The present invention utilizes the SLAF-seq technology to carry out gene sequencing on the sea urchin mesenterus, and obtains a large number of SLAF tags in the sequencing, and the SLAF tags can be used to construct a high-density genetic map, and utilizes the genetic map and the phenotypic traits of the sea urchin mesenterus (shell height, shell diameter) , body weight) for association analysis and QTL mapping, through the QTL screening of the target traits (shell height, shell diameter, body weight), the SLAF tags related to growth traits (shell height, shell diameter, body weight) can be deduced back. Among them, SLAF-seq technology and HighMap software were used to develop and construct genetic maps, and the interval mapping (IM) algorithm of MapQTL software and the composite interval mapping (CIM) algorithm of RQTL software were used for QTL positioning. Specifically, for the method of constructing the genetic map of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com