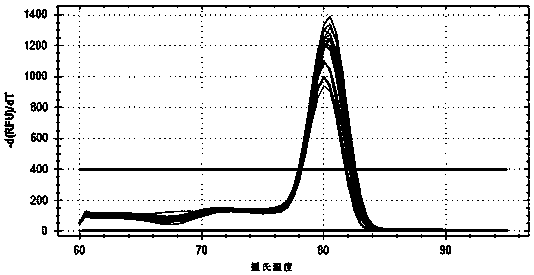
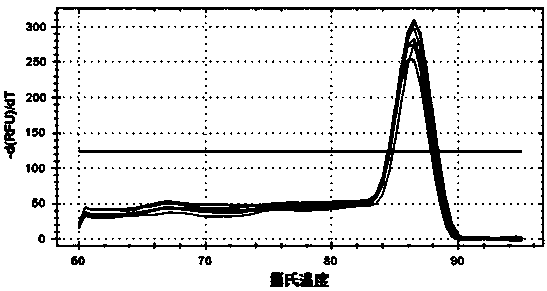
Primers, kit and method to detect transcriptional level of purine nucleotide phosphorylase PNP gene in macaque liver by RT-qPCR (real-time quantitative polymerase chain reaction)
A purine nucleoside phosphatase and gene transcription technology, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., to achieve high sensitivity, high repeatability, and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Primers for RT-qPCR detection of liver purine nucleoside phosphatase PNP gene transcription levels in macaques, including macaques PNP Specific upstream and downstream primer pairs for gene expression levels and specific upstream and downstream primer pairs for macaque GAPDH gene as an internal reference gene;
[0060] Among them, macaques PNP The specific upstream and downstream primers for gene expression level are:
[0061] PNP F: 5'-gagaccatggagaacggatacac-3'
[0062] PNP R: 5'-cagacctcctaatccagaaccac-3'
[0063] The specific upstream and downstream primers of the rhesus monkey GAPDH gene as an internal reference gene are:
[0064] GAPDH F: 5'-agccccatcaccatcttcc-3'
[0065] GAPDH R: 5'-aatgagccccagccttctc-3'.
Embodiment 2
[0067] A kit for detecting the transcription level of purine nucleoside phosphatase PNP gene in rhesus monkey liver, comprising the primers described in Example 1, SYBR Premix Ex Taq II enzyme, cDNA template of rhesus monkey liver tissue and deionized water.
Embodiment 3
[0069] A method for RT-qPCR detection of macaque liver purine nucleoside phosphatase PNP gene transcription level, comprising the following steps:
[0070] (1) Design the following primers:
[0071] According to the macaque in the NCBI gene bank ( macaca mulatta )of PNP Nucleotide sequence number: NM_001193551, rhesus monkey ( macaca mulatta )of GAPDH Nucleotide sequence number NM_001195426.1 designed primers, the above primers were synthesized by Shanghai Bioengineering Co., Ltd.:
[0072] Macaque PNP The specific upstream and downstream primers for gene expression level are:
[0073] PNP F: 5'- gagaccatggagaacggatacac-3'; (SEQ ID NO.1)
[0074] PNP R: 5'-cagacctcctaatccagaaccac-3'; (SEQ ID NO.2)
[0075] The specific upstream and downstream primers of the rhesus monkey GAPDH gene as an internal reference gene are:
[0076] GAPDH F: 5'-agccccatcaccatcttcc-3'; (SEQ ID NO.3)
[0077] GAPDH R: 5'-aatgagccccagccttctc-3'. (SEQ ID NO.4)
[0078] Macaque ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com