A method for screening SNP sites used for detecting sample contamination in high-throughput sequencing and a sample contamination detecting method
A screening method and high-throughput technology, which can be used in biochemical equipment and methods, and the determination/inspection of microorganisms.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0030] According to a typical implementation of the present invention, it is mainly divided into two parts:
[0031] Part 1: Sample Processing
[0032] It mainly includes: breaking DNA, adding adapters, hybridization capture, elution, enrichment, and sequencing.
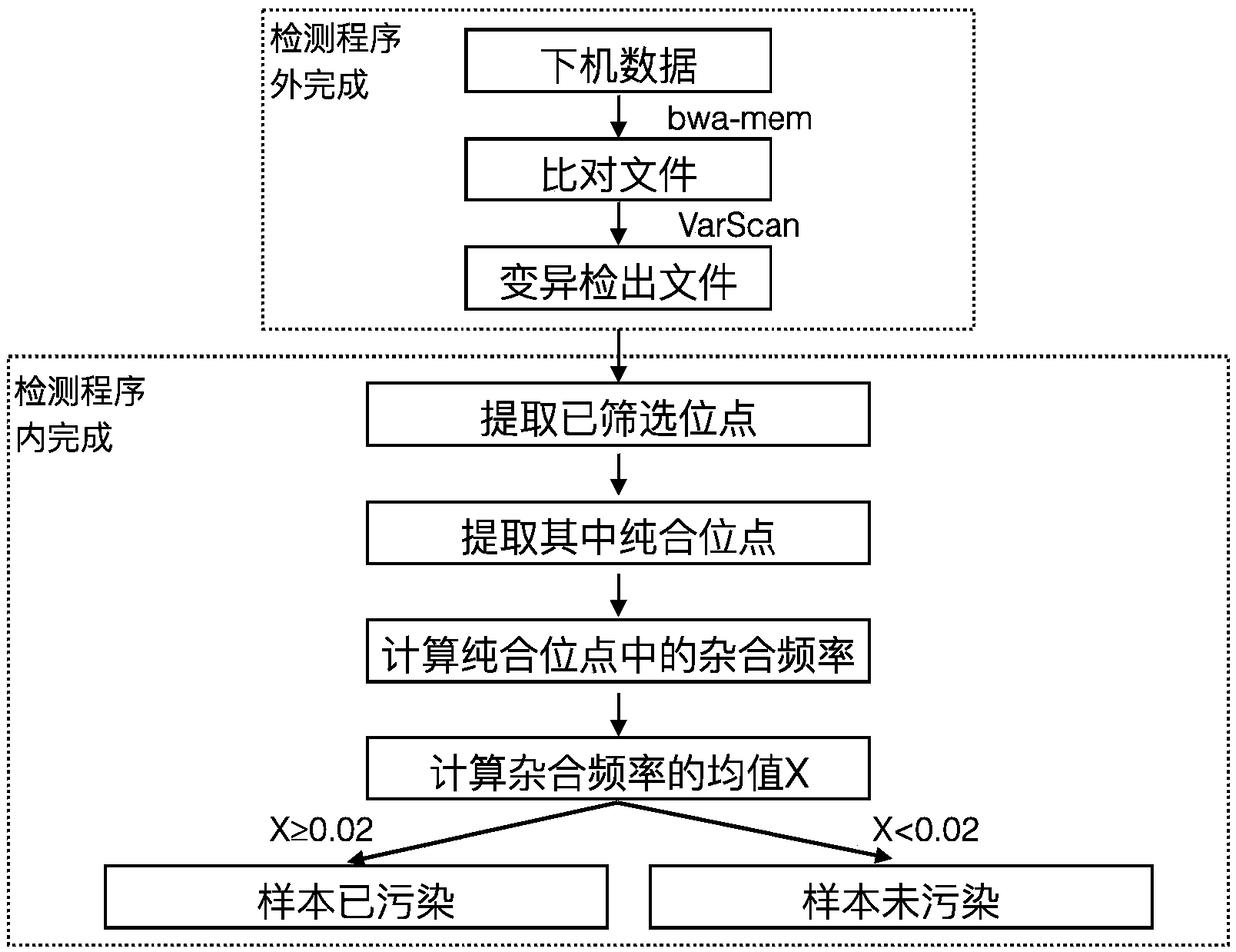
[0033] Part II: Data processing, such as figure 1 The flow shown includes two parts: completion outside the detection procedure and completion within the detection procedure. Among them, the completion of the detection program mainly includes: use the BWA-mem comparison software to compare the high-throughput sequencing sequence to the human reference gene on the off-machine data, and the unmatched sequence forms a soft truncation; and then according to the position of the comparison. Sorting, and using samtools software to create an index; using VarScan software to input the compared files for mutation detection. The completion of the detection program mainly includes: extracting the screened sites, determining t...
Embodiment 1
[0036] SNP site screening includes:
[0037] 1) Select the SNP sites in the 1000Genome Project project, which needs to satisfy 40%<population frequency<60%, a total of 35824769;
[0038] 2) To select the SNP sites in the HapMap project, 40%
[0039] 3) Select the SNP sites shared by the 1000Genome Project and the HapMap project, a total of 1,086,179;
[0040] 4) Screen the consensus sites covered by the probes used to capture the target region during the sequencing process, a total of 7554;
[0041] 5) Design probes for the above sites and perform sequence alignment with the Hg19 human reference genome, remove sites with more than 1 compared position, and the remaining 7328;
[0042] 6) The 7328 sites are sorted according to the absolute position of the chromosome, and extracted with a step size of 2;
[0043] 7) Finally, 3664 SNP sites evenly distributed in the Panel were obtained for subsequent evaluation. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com