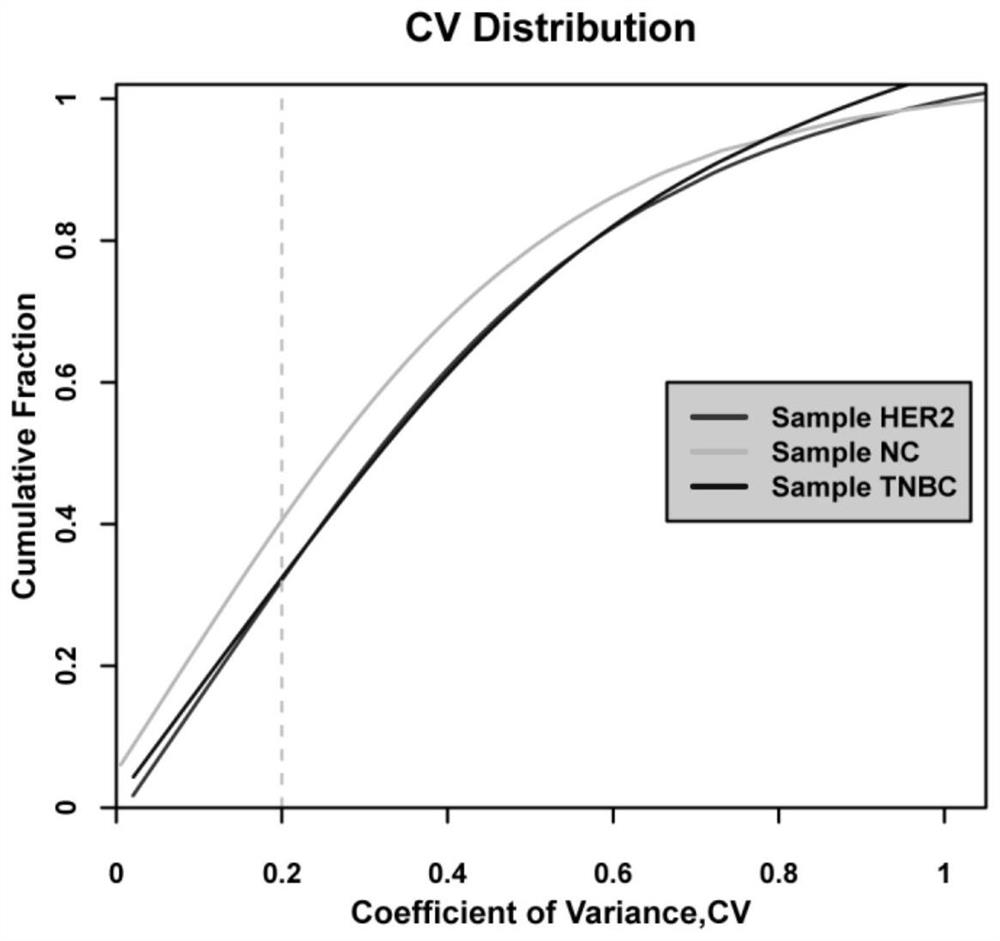
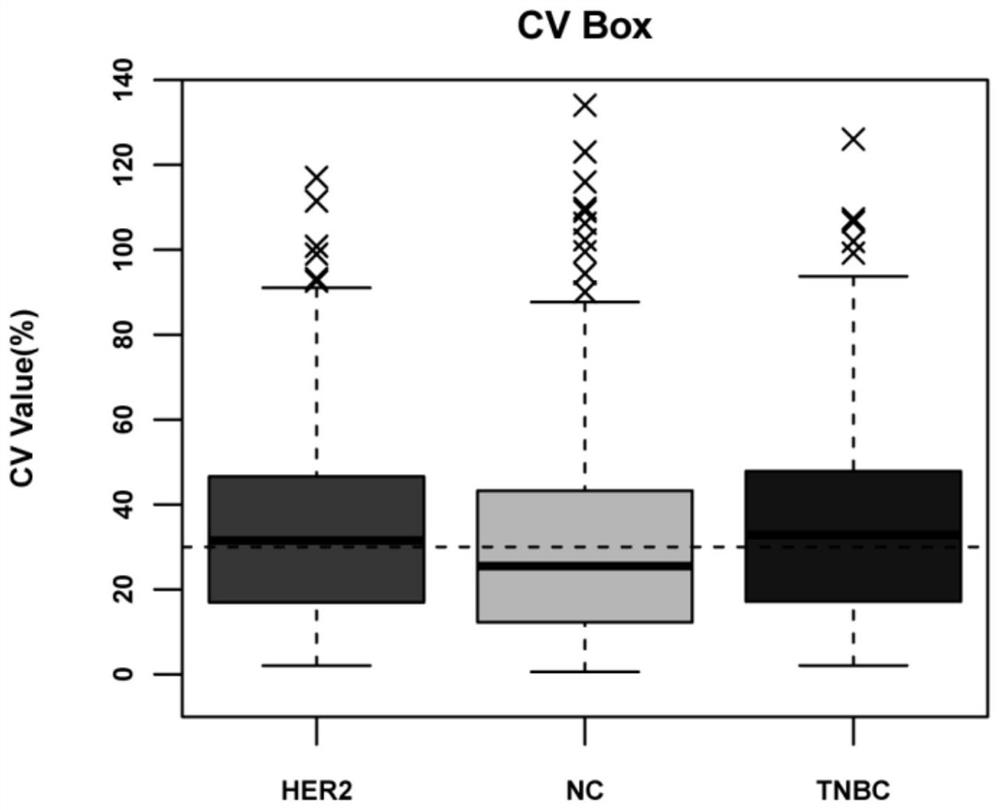
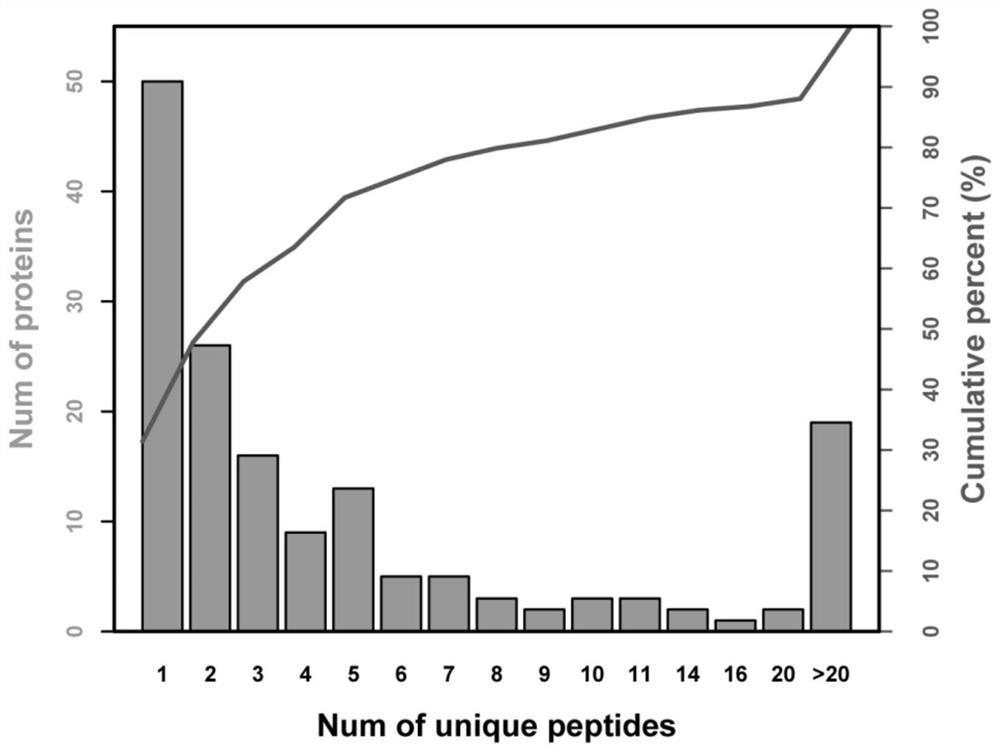
Application of itraq technology to study the method of differentially expressed proteins in exosomes of triple-negative breast cancer
A technology for triple-negative breast cancer and differentially expressed proteins, applied in instruments, measuring devices, scientific instruments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0045] The present invention will be described in further detail below in conjunction with the accompanying drawings and specific embodiments.
[0046] In the embodiment of the present invention, the present invention provides a method of applying iTRAQ technology to study differentially expressed proteins in triple-negative breast cancer exosomes.
[0047] Unless otherwise specified in the examples of the present invention, all reagents and consumables used are commercially available.
[0048] 1 Materials and methods
[0049] 1.1 Sample preparation (model building)
[0050] Three groups of blood samples, among them, the experimental group A is the blood samples of patients with triple-negative breast cancer, the control group B is the blood samples of HER2-positive breast cancer patients, and the control group C is the blood samples of normal people, in which the experimental group A has three samples, and the control group Three samples of group B, two samples of control g...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com