Methods and systems of molecular recording by crispr-cas system
A DNA sequence and cell technology, applied in biochemical equipment and methods, DNA/RNA fragments, DNA preparation, etc., can solve problems such as lack and limitation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Materials and methods
[0078] Bacterial Strains and Culture Conditions
[0079] Expression and neospacer collection were performed in BL21-AI cells. Cells were grown at 37°C in Luria Broth (LB) with shaking (240 rpm) unless otherwise stated. L-arabinose (Sigma-Aldrich) (from 20% stock solution in water) was used at 0.2% final concentration (w / w) and isopropyl-β-D-sulfur at 1 mM final concentration Substituted galactopyranoside (IPTG; Sigma-Aldrich) (from a 100 mM stock in water) induced gene expression from the T71ac promoter. Cas mutants expressed from the pLtetO promoter were induced by anhydrotetracycline (aTc; Clontech) at a final concentration of 214 nM (from a 214 μM stock solution in 50% ethanol). Concurrently with expression from the pLtetO promoter, 0.2% glucose was added to reduce unintended background expression from the T71ac promoter. For new spacer harvesting experiments not involving oligonucleotide-derived spacers, cells were induced and grown overn...
Embodiment 2
[0089] Type I-E CRISPR-Cas systems accept synthetic spacers in vivo.
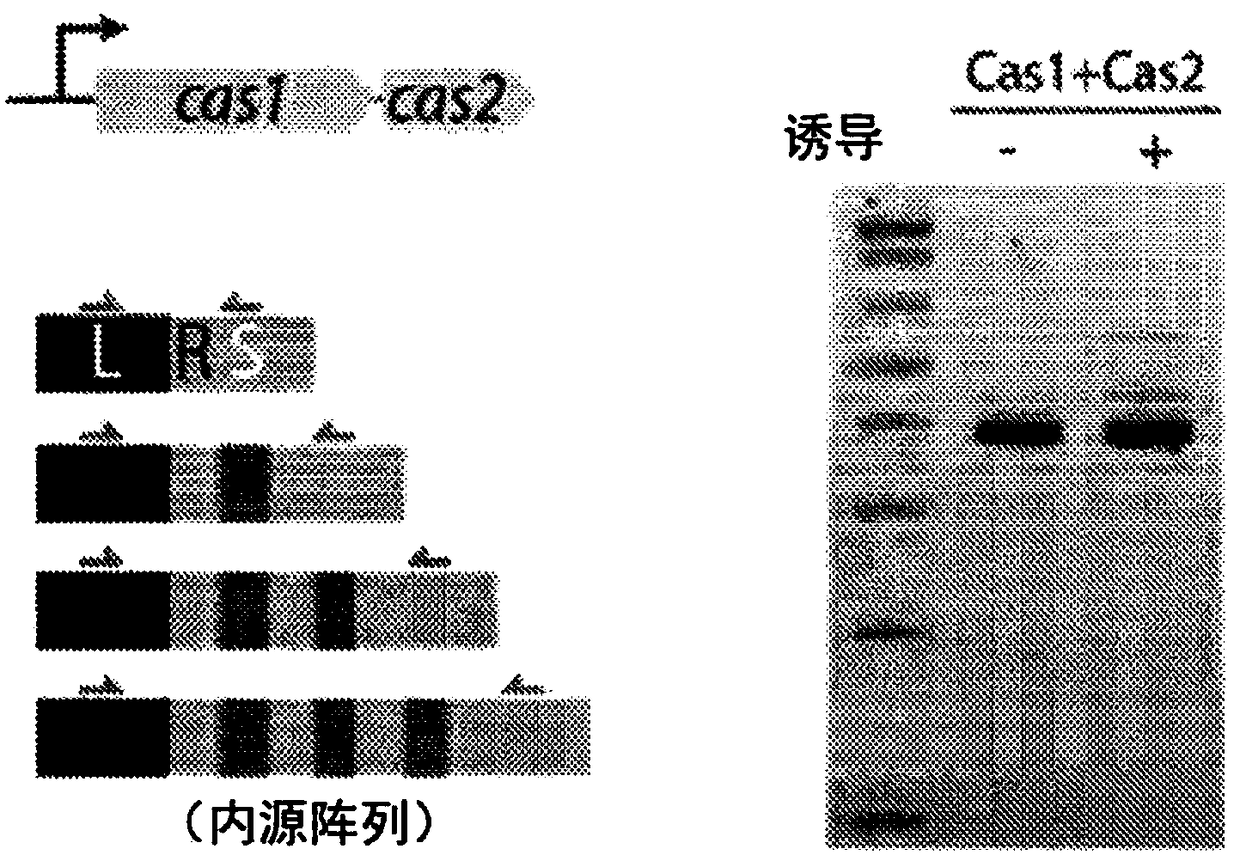
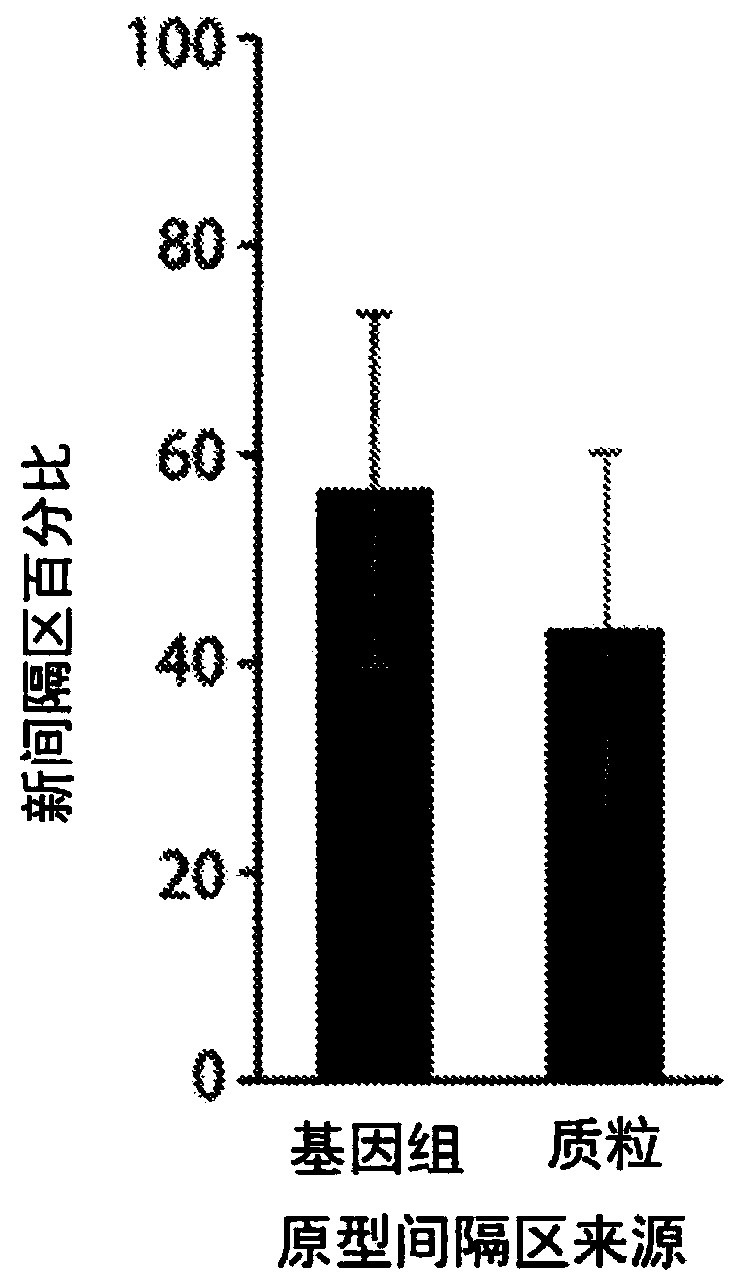
[0090] It was recently shown that overexpression of the Escherichia coli type I-E CRISPR-Cas proteins Cas1 and Cas2 is sufficient to drive acquisition of new spacers in a strain (BL21-AI) containing two genomic CRISPR arrays but lacking endogenous Cas proteins (see I. Yosef , M.G.Goren, U.Qimron, Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic acids research 40, 5569-5576 (2012); Epub online publication , July (10.1093 / nar / gks216)). The result is replicated (see Figure 1A ), and similarly a new spacer was found to be consistently integrated into the first position of Array I, which is directly adjacent to the leader, with a consistent size of 33 bases (see Figure 2A -B). Roughly equal numbers of these spacers were extracted from the cell's own genome and from the plasmids used to overexpress Cas1 and Cas2 (see Figure 1B ). Considering the overall DN...
Embodiment 3
[0095] PAM changes the efficiency and directionality of spacer acquisition.
[0096] Using data collected by our sequencing of millions of arrays after amplification, it was found that genomic and plasmid-derived protospacers were overall drawn in equal numbers from the forward and reverse strands, with the only apparent overall bias pointing towards genome duplication starting point (see Figure 3A ). Similarly, oligonucleotide-derived protospacers were also found to have equal proportions in the forward and reverse directions in the array (see Figure 3B ). When the context of the genomic and plasmid-derived protospacer was further examined, strong evidence for a PAM at the 5' end of the protospacer was found, consisting of two adenines from positions -2 and -1 of the spacer, and Strong preference for guanine as the first spacer base (see Figure 3C ). This is largely consistent with previous characterization of E. coli type I-E systems (see I. Yosef, D. Shitrit, M.G. G...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com