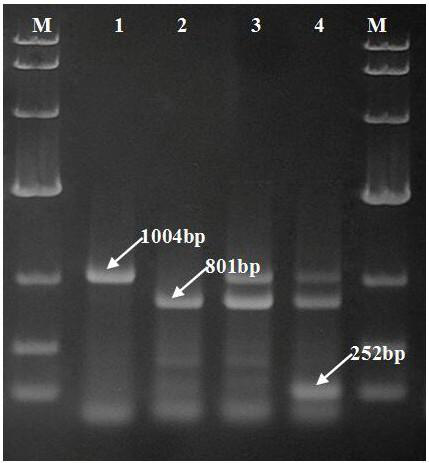
A SSR-PCR method for identification of four main cultivars of Agaricus bisporus
The technology of a main species, Agaricus bisporus, is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., and can solve the problem of irregular multiplication and species conservation, same name and different species, and same species with different names and other problems, to achieve the effect of time-saving and labor-saving identification, easy operation and accurate identification
Active Publication Date: 2019-01-15
INST OF EDIBLE FUNGI FUJIAN ACAD OF AGRI SCI
View PDF3 Cites 2 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
However, because Agaricus bisporus can reproduce asexually, some farmers and enterprise growers often separate through organizations, multiply and preserve species irregularly, resulting in the phenomenon of the same species with different names and different species with the same name in the process of production, management and circulation. , so it is very necessary to establish a set of rapid identification methods for domestic main varieties
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment 1
[0016] 1. Taq DNA polymerase, PCR buffer, MgCl 2 , dNTPs were purchased from Xiamen Taijing Biological Co., Ltd., and the primers were synthesized by Sangon Bioengineering (Shanghai) Co., Ltd., and the PCR product sequence was sequenced by Sangon Bioengineering (Shanghai) Co., Ltd.
[0017] The nucleotide sequence of the specific primer is:
[0018] Scaf4_1k11F: 5'-CAATCACAATCCCTCCAAGC-3',
[0019] Scaf4_1k11R: 5'-AAACGAATTAGGAACGGTGG-3';
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
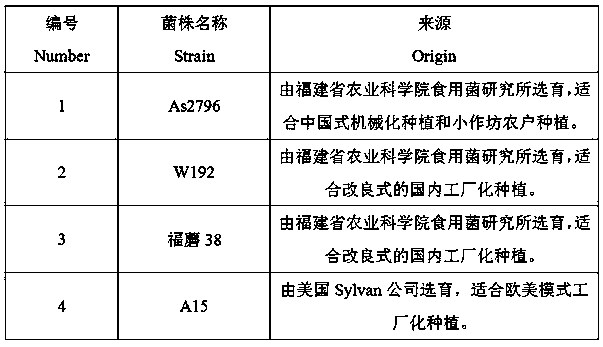
The invention belongs to the field of molecular marker assisted breeding of edible fungi, in particular to an SSR method for identification of four main cultivars of Agaricus bisporus. In this method, SSR markers are used to amplify the PCR products of As2796, W192. Tricholoma fumefaciens 38 and A15, which are the main cultivars of Agaricus bisporus in China. The method of the invention does notrequire a miscellaneous agronomic character mushroom production comparison test, the specific amplified map can identify the main cultivars of Agaricus bisporus in China. This method can be used to identify 4 main cultivars of Agaricus bisporus in China quickly, avoiding synonym and synonym phenomenon, and improving the production efficiency of Agaricus bisporus.
Description
technical field [0001] The invention belongs to the field of molecular marker assisted breeding of edible fungi, and relates to an SSR-PCR identification method for distinguishing four main cultivars of Agaricus bisporus As2796, W192, Fumo 38 and A15. Background technique [0002] Simple sequence repeat (SSR), also known as microsatellite DNA, is a tandem repeat sequence with 2-6 bases as the repeating motif, which widely exists in the genomes of higher organisms. Among the many molecular marker methods, genome-based SSR markers have the characteristics of numerous specific sites, co-dominance, multiple alleles, and wide distribution. At present, they have become the most important methods for constructing linkage genetic maps, studying population genetics, drawing breed fingerprints, and so on. It is an ideal tool for screening individual specific bands and molecular marker-assisted breeding. [0003] Agaricus bisporus ( Agaricus bisporus ) is the most widely cultivated e...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More IPC IPC(8): C12Q1/6895C12Q1/04C12N15/11
CPCC12Q1/6895C12Q2600/156C12Q2600/16
Inventor 蔡志欣陈美元廖剑华郭仲杰卢园萍施肖堃王泽生
Owner INST OF EDIBLE FUNGI FUJIAN ACAD OF AGRI SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com