Method and system for predicting association relationship between disease and LncRNA
A related relationship and disease technology, applied in the field of bioinformatics disease prediction, can solve the problems of high time and cost of biological experiments, achieve the effect of reducing the cost and overhead of biological experiments and accelerating research progress
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
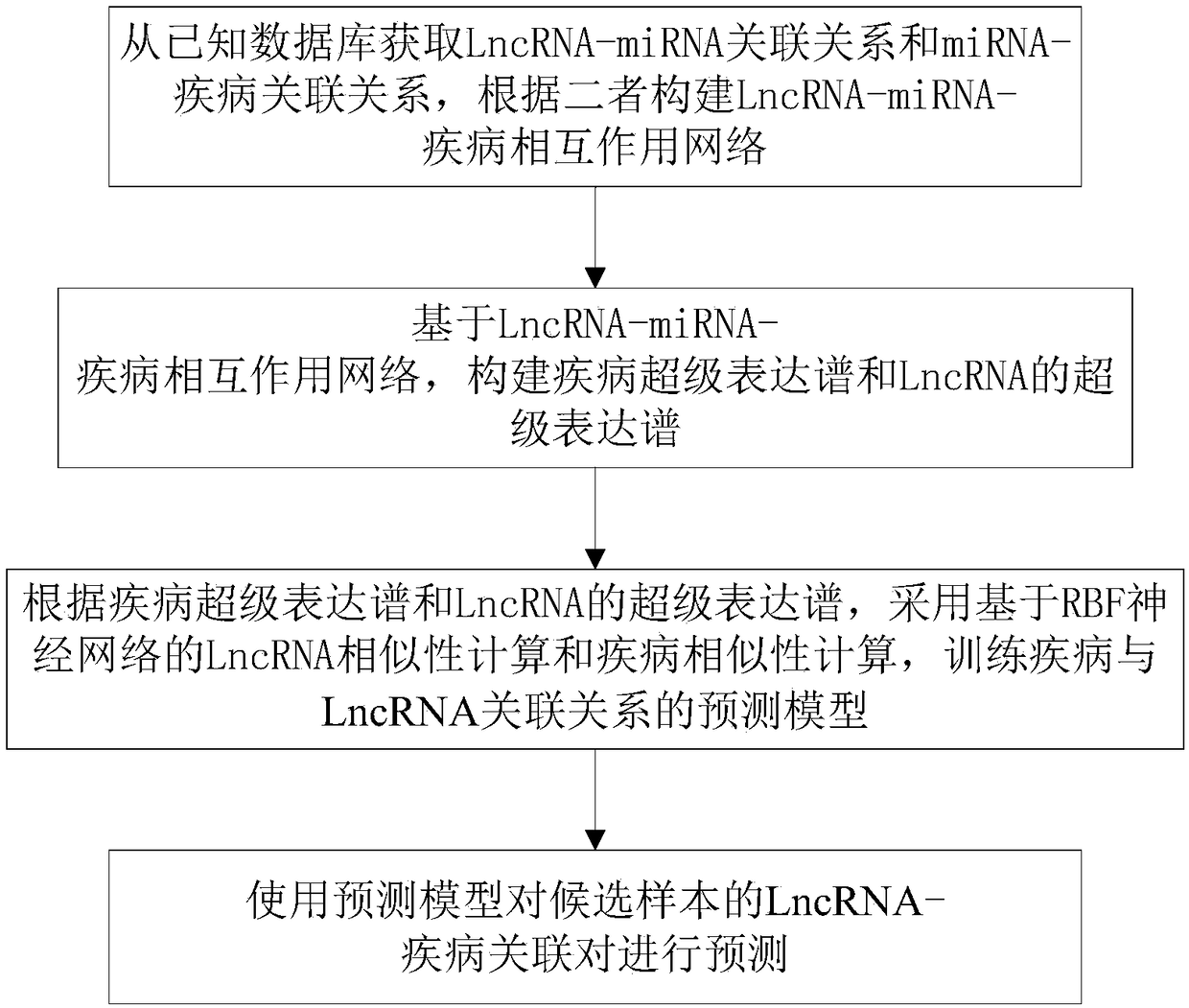
[0056] The method for predicting the relationship between disease and LncRNA in this embodiment comprises the following steps:
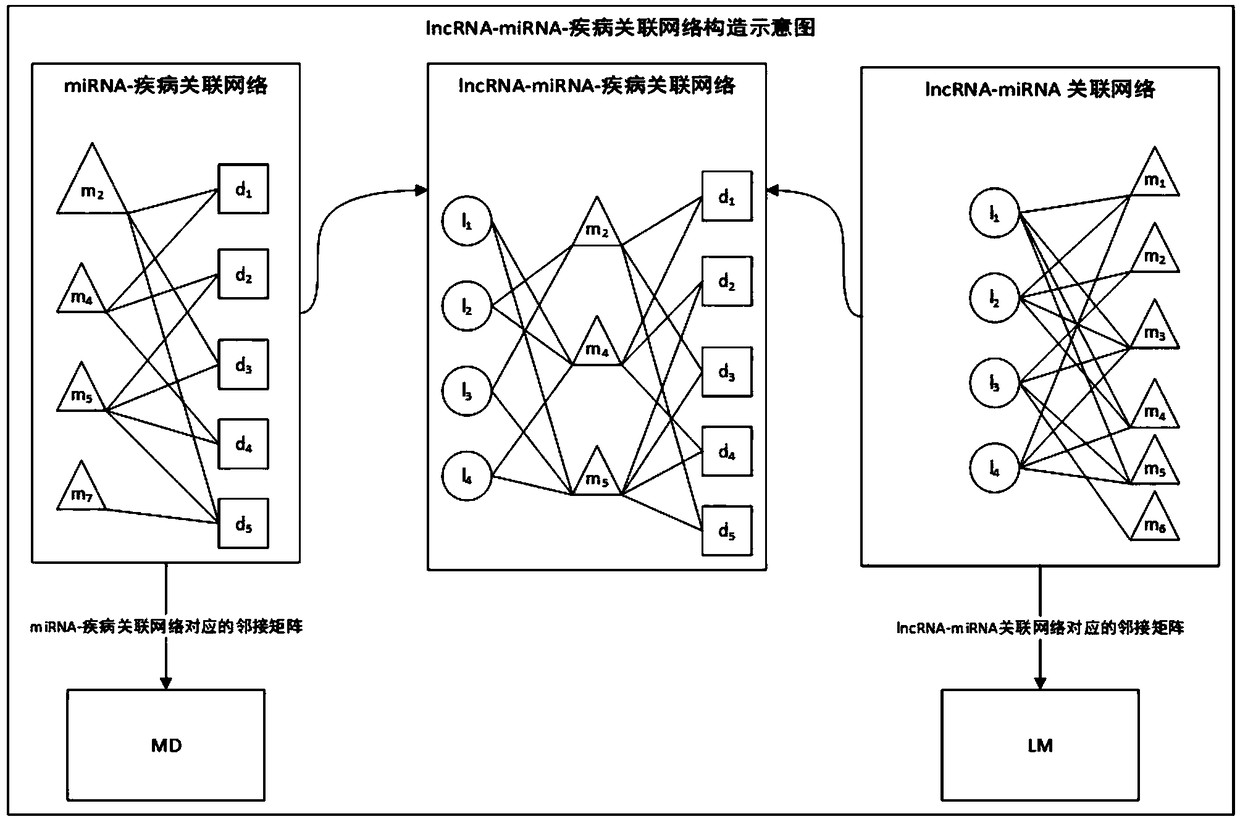
[0057] S1: Obtain the LncRNA-miRNA association relationship and miRNA-disease association relationship from known databases, and construct the LncRNA-miRNA-disease interaction network based on the two. details as follows:
[0058] S11: Obtain the LncRNA-miRNA association relationship data set from the known database, and obtain the LncRNA-miRNA interaction pair;
[0059] S12: Obtain miRNA-disease association data sets from known databases, and obtain miRNA-disease interaction pairs;
[0060] S13: Construct a LncRNA-miRNA association network based on LncRNA-miRNA interaction pairs;
[0061] S14: Construct a miRNA-disease association network based on miRNA-disease interaction pairs;
[0062] S15: Based on the LncRNA-miRNA association network and the miRNA-disease association network constructed above, construct an LncRNA-miRNA-disease interaction ne...
Embodiment 2
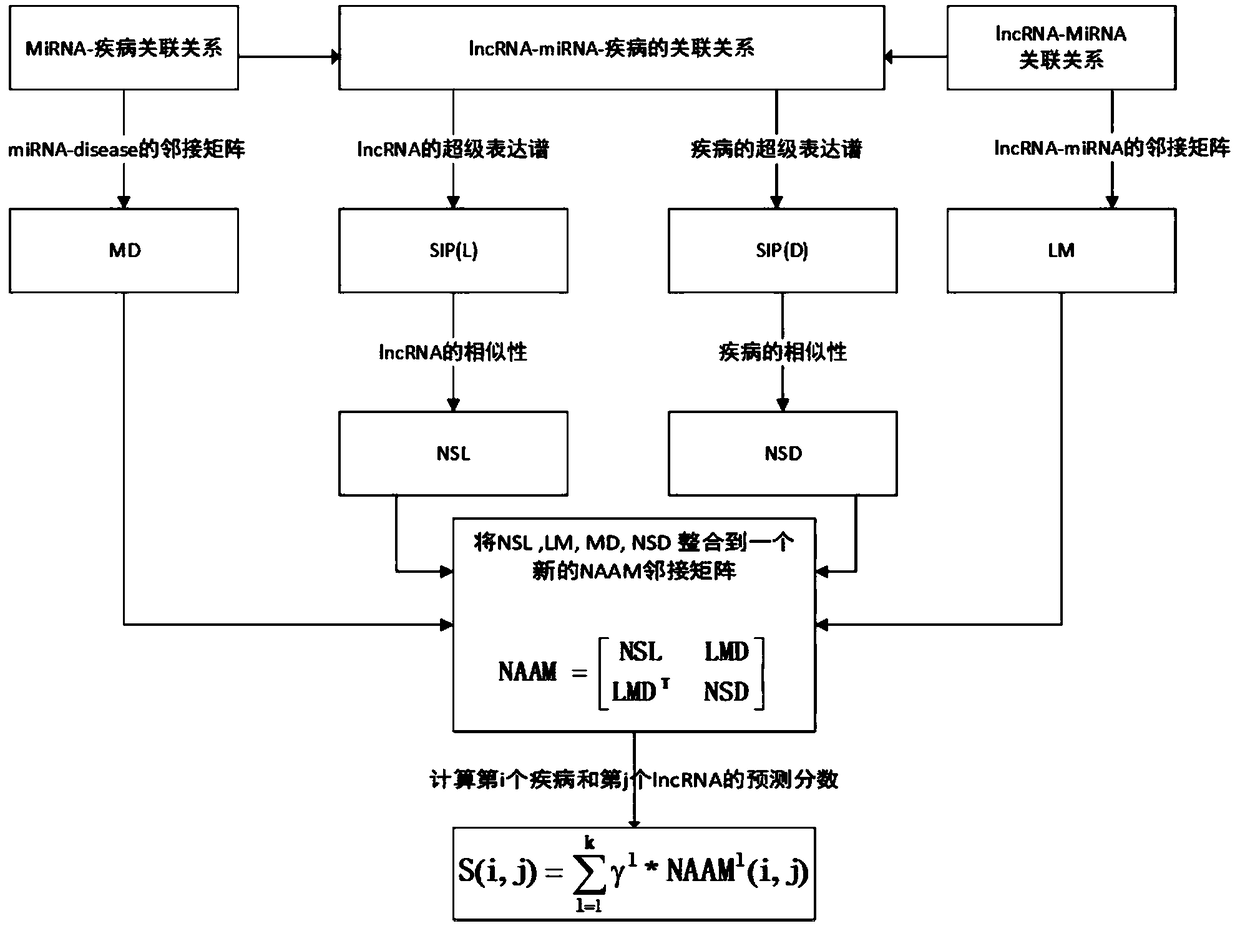
[0078] figure 2 Is the overall flowchart of the present embodiment, wherein, MD is the adjacency matrix of miRNA-disease, LM is the adjacency matrix of LncRNA-miRNA, SIP (L) is the super expression spectrum matrix of LncRNA, SIP (D) is the super expression of disease Spectrum matrix, NSL is the similarity matrix of LncRNA, NSD is the similarity matrix of disease, LMD=LM*MD, LMD T is the transpose of the LMD matrix, and S(i,j) is the prediction score of the i-th disease and the j-th LncRNA.
[0079] This embodiment provides a method for predicting the relationship between disease and LncRNA based on the LncRNA-miRNA-disease interaction network, including the following steps:
[0080] S1: Acquisition and processing of raw data sets, preprocessing and feature selection of data features. details as follows:
[0081] S11: Acquisition of LncRNA-miRNA association relationship;
[0082] Two versions of the LncRNA-miRNA association dataset (version 2015 and version 2017) were down...
Embodiment 3
[0154] As a general technical concept, this embodiment also provides a system for predicting the relationship between diseases and LncRNAs corresponding to the methods in the above embodiments, including: a network construction unit for obtaining LncRNA-miRNA associations from known databases Relationship and miRNA-disease association relationship, construct LncRNA-miRNA-disease interaction network according to the two; expression profile construction unit, used to construct disease super-expression profile and LncRNA super-expression based on the LncRNA-miRNA-disease interaction network Spectrum; Model construction unit, for according to the super expression profile of described disease and LncRNA, adopt the LncRNA similarity calculation and disease similarity calculation based on RBF neural network, train the prediction model of disease and LncRNA correlation; Prediction unit , for predicting LncRNA-disease association pairs of candidate samples using the predictive model.
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com