Leon-RC compression method for genome sequencing data
A genome sequencing and compression method technology, applied in the field of biological information, can solve the problems of low compression rate, long time to find anchor points, no consideration of mirror repetition, inversion repetition, etc., to reduce size, optimize construction process, reduce effect of size
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
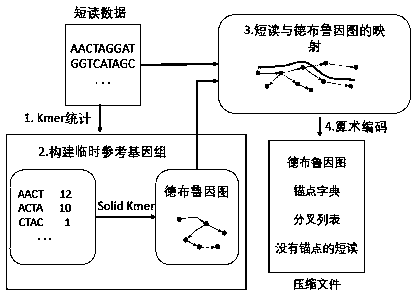
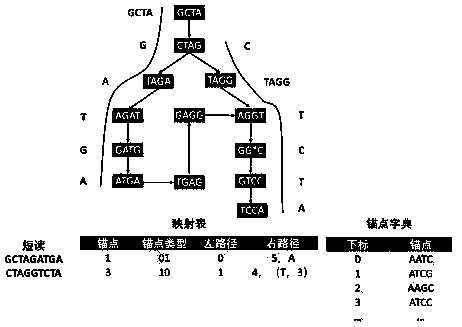
[0038] The present invention provides a Leon-RC compression method of genome sequencing data, which mainly improves the steps of constructing anchor point dictionary by LEON algorithm, including the following steps:
[0039] (1) Divide short reads into multiple Kmers;
[0040] (2) Select a Kmer, calculate the Kmer value of its direct repetition, mirror repetition, inverted repetition, and complementary palindrome, compare these four values, and obtain the smallest Kmer value;
[0041] (3) Put the smallest Kmer value into the Bloom filter for matching search. SolidKmer is stored in the Bloom filter, and judge whether there is the smallest Kmer value in the Solid Kmer; if it exists, add it to the anchor dictionary The smallest Kmer value, and end the search; if it does not exist, get the next Kmer, repeat steps (2), (3);
[0042] (4) If the smallest Kmer value of all Kmers does not exist in Solid Kmer, it means that there is no anchor point for the short read;
[0043] (5) Con...
Embodiment 2
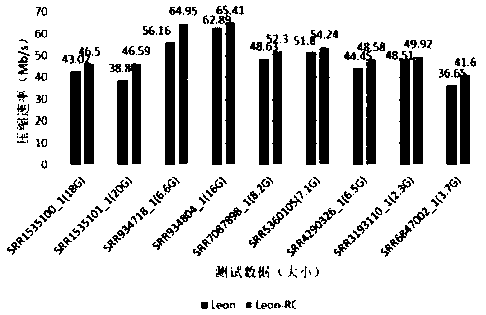
[0058] In this embodiment, compression tests are performed on next-generation sequencing data of different sizes, and the results of the compression tests are as follows image 3 As shown, compared with Leon, Leon-RC significantly improves the compression rate while keeping the compression rate unchanged. Among them, the compression rate of the SRR934718_1 file has increased the most, from 56.16Mb / s to 64.95Mb / s. The increase rate is as high as 15.6%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com