SNP fluorescence in situ hybridization sequencing detection method for ABCB1 and SLCO1B1
A fluorescence in situ hybridization and detection method technology, applied in the field of molecular genetics, can solve the problems of death, high misjudgment rate, high chip cost, etc., and achieves the effects of high identification, not easy to contaminate, and fast sequencing speed.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0063] The present invention will be further described below in conjunction with specific embodiments, and the advantages and characteristics of the present invention will become clearer along with the description. However, these embodiments are only exemplary and do not constitute any limitation to the scope of the present invention.
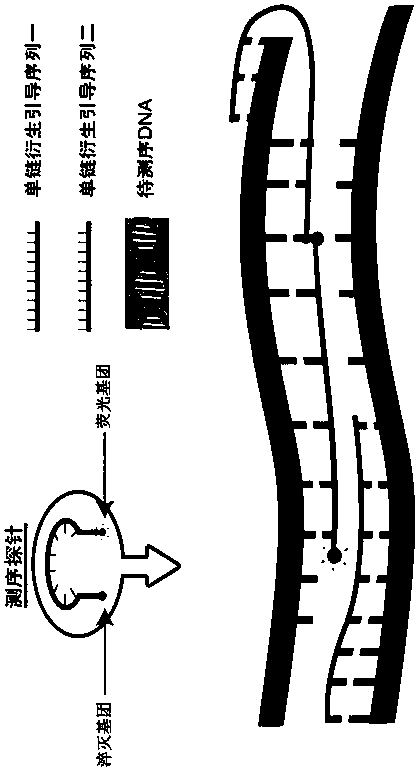
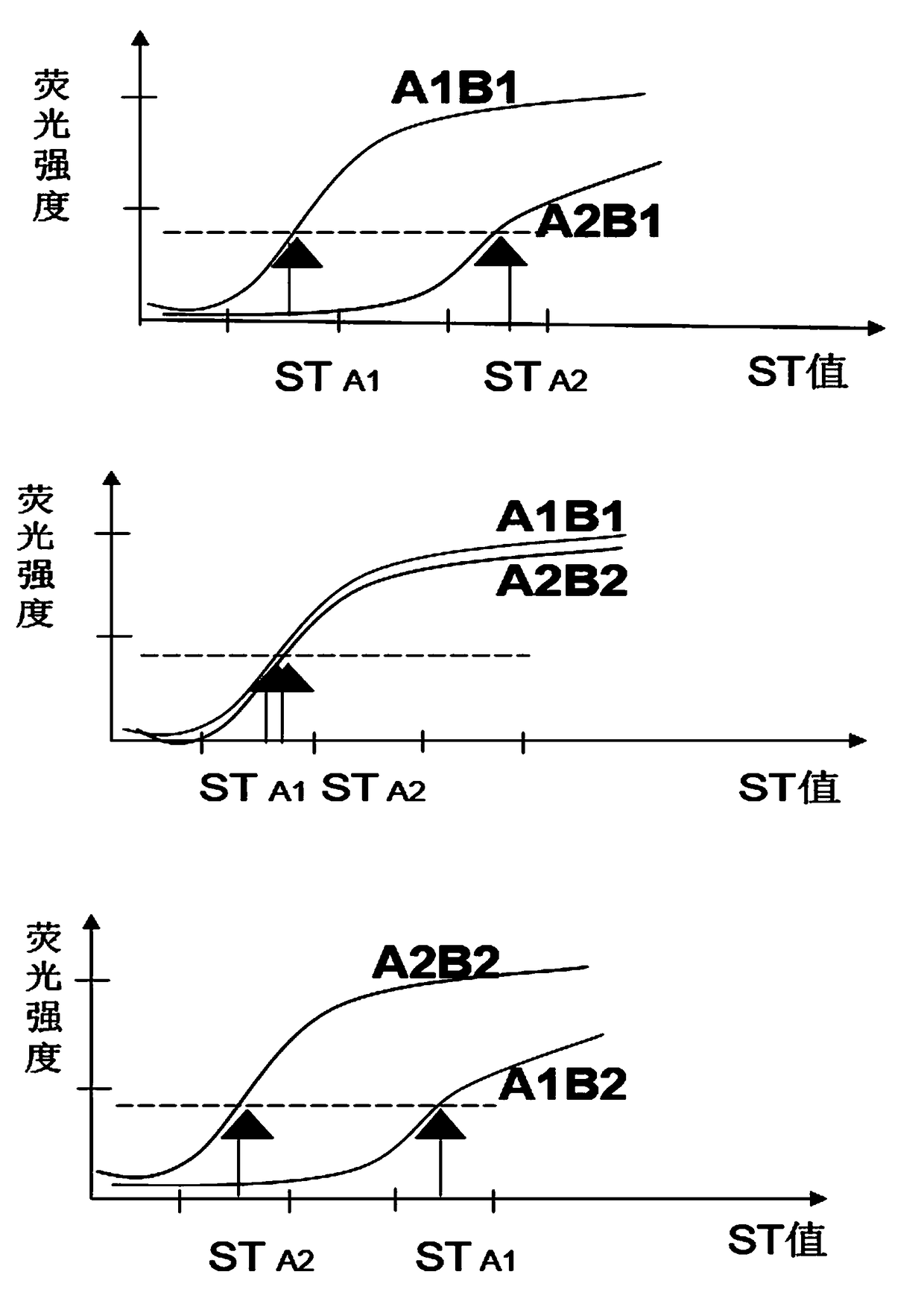
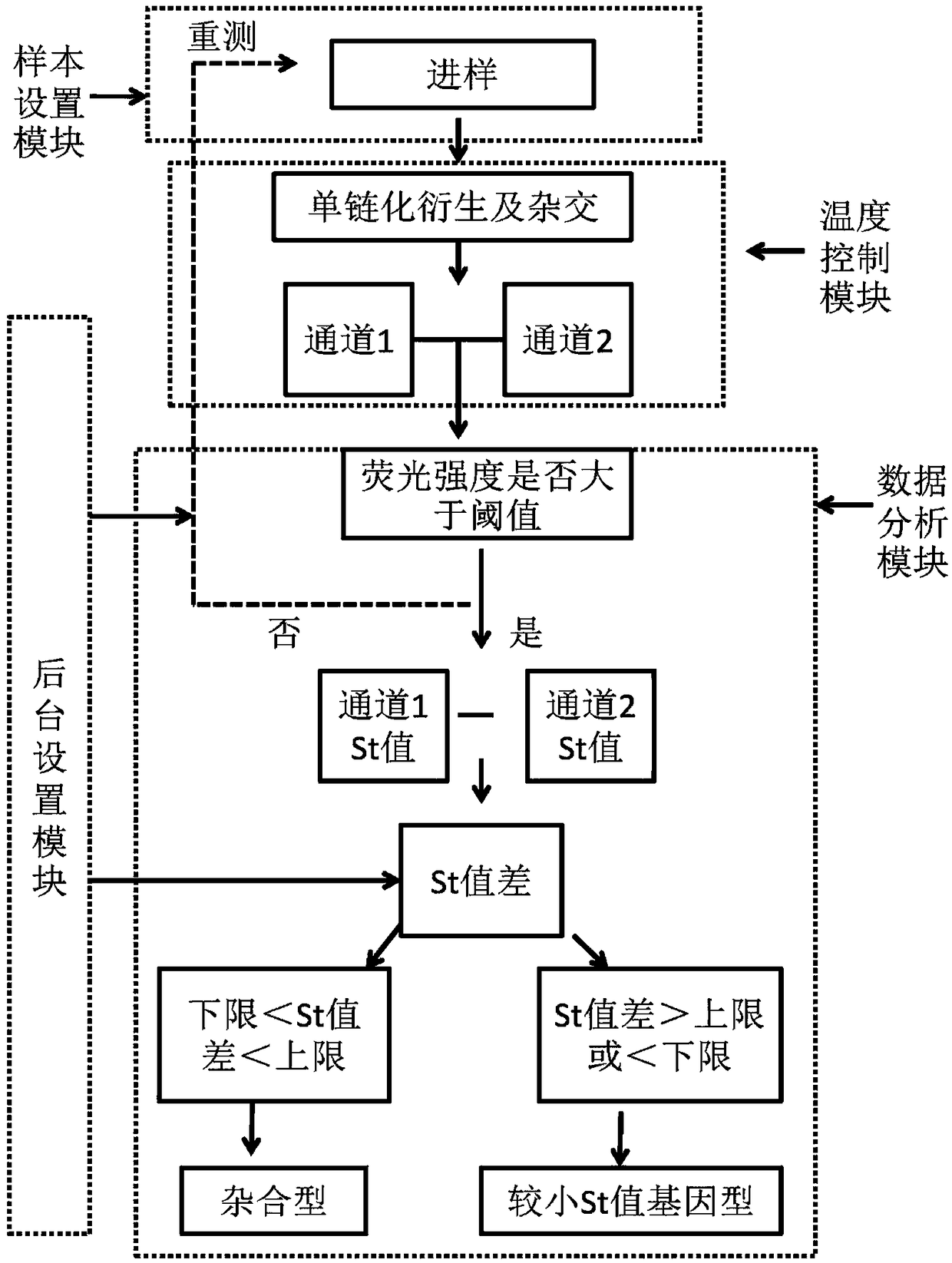
[0064] Reaction principle
[0065] Different from ordinary Taqman technology, the present invention does not use PCR technology in the preparation of hybridization templates, but uses single-stranded derivation guide oligonucleotide sequences to convert DNA double-stranded templates into single-stranded derivatives, and Direct hybridization with labeled probes for sequencing.
[0066] The single-stranded derivation requires an oligonucleotide guide sequence capable of complementary binding to the template strand to initiate, and the guide sequence can specifically bind to the annealed template strand at the 3' end and / or 5' end of the template...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com