DNA sequence optimization method based on particle swarm chaotic intrusion weed algorithm
An optimization method and coding sequence technology, applied in the fields of genomics, instrumentation, proteomics, etc., can solve problems such as incorrect calculation, and achieve the effect of good global search ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
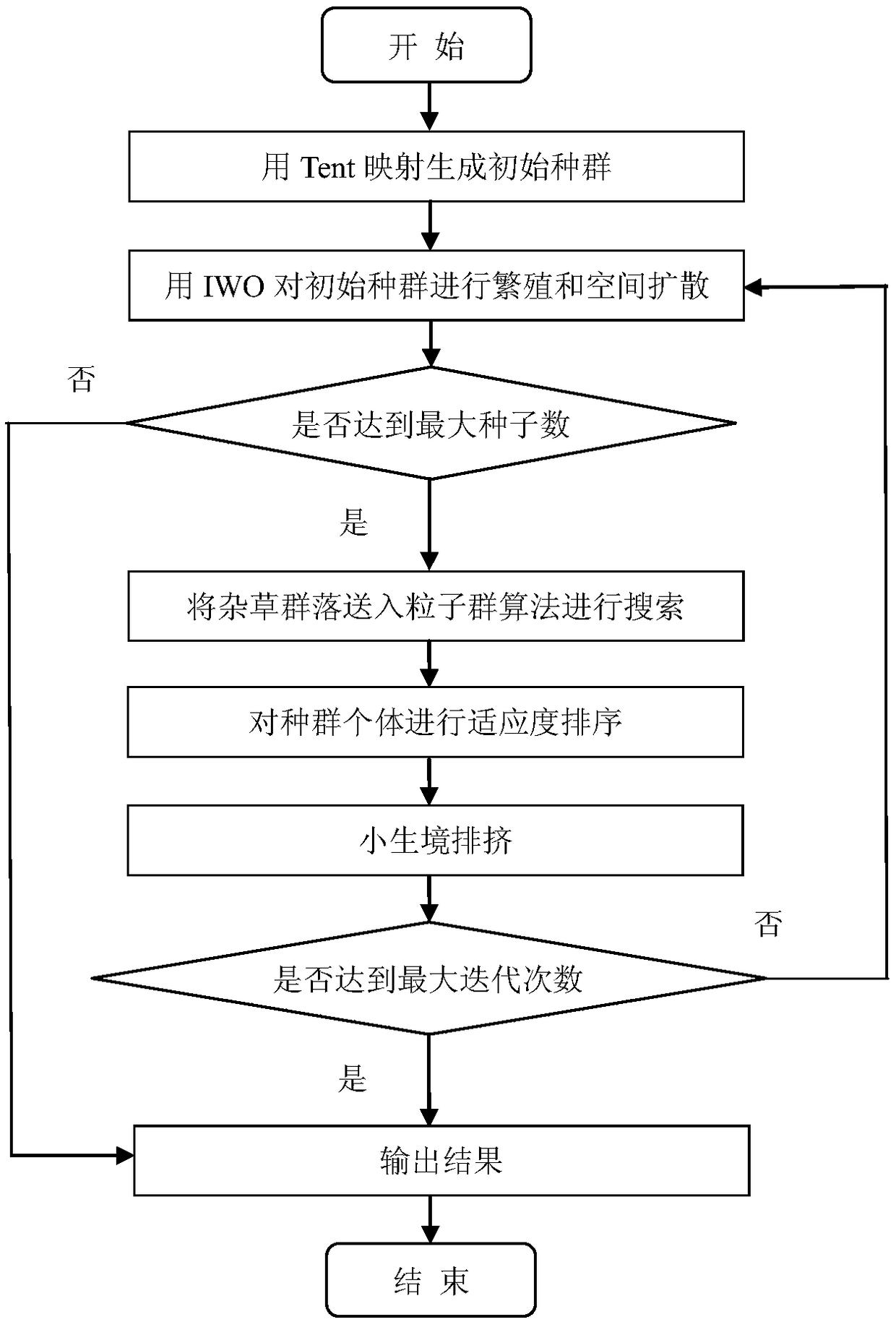
[0021] Such as figure 1 As shown, the realization flowchart of the DNA sequence optimization method based on the particle swarm-invading weed algorithm of chaos, the method includes the following steps:
[0022] Step 1: Use Tent mapping to map the random initial population, and the obtained population is used as the initial population;
[0023] The initial DAN sequence is shown in Table 1.
[0024] Table 1:
[0025]
[0026] Step 2: Use the invasive weed algorithm to evolve the initial population, calculate the number of seeds that each population individual can generate, multiply these seeds into weeds, and distribute the weeds generated by the seeds among the parent individuals in a Cauchy distribution manner Around, calculate the fitness value of the merged offspring individuals and parent individuals;
[0027] Step 3: Compare the number of weeds obtained by the evolution of the invasive weed algorithm with the preset maximum number of weeds, if the number is greater ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com