Method for in vivo seamless assembly of DNA
An assembly method, seamless technology, applied in the field of molecular biology, can solve the problem of long time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Example 1, primers and method design for splicing crtE, crtB and crtI three genes
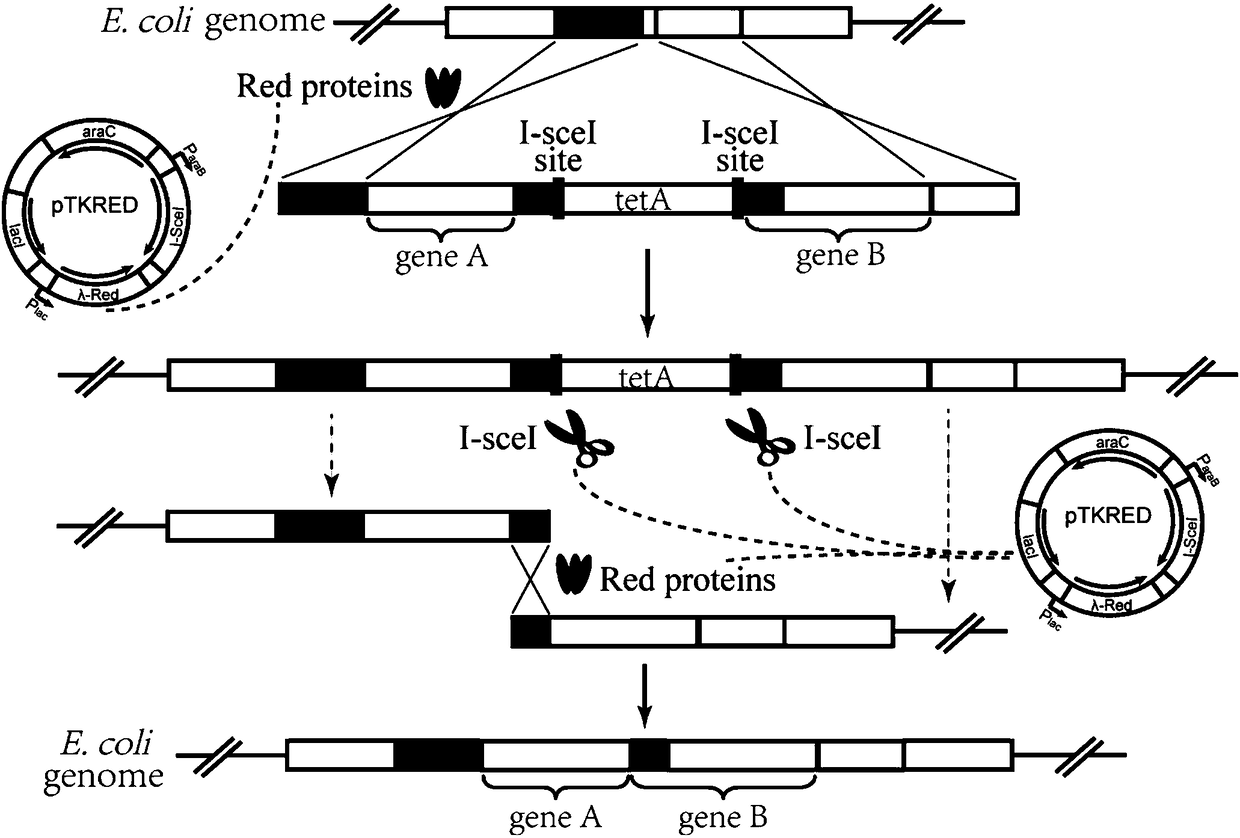
[0071] In this embodiment, three genes of crtE, crtB and crtI are seamlessly spliced as an example. Positive clones were selected by tetA gene negative screening technique. First use crtE as gene A and crtB as gene B.
[0072] In the present embodiment, the DNA sequence of each gene involved is as follows:
[0073] (1) The promoter sequence used by the tetA gene (SEQ ID NO: 1):
[0074] ATCG TTGACG GCTAGCTCAGTCCTAGG TACAGT GCTAGCTACTAGAGAAAGAGGAGAAATACTAG
[0075] (2) tetA gene sequence (SEQ ID NO: 2):
[0076]ATGAATAGTTCGACAAAGATCGCATTGGTAATTACGTTACTCGATGCCATGGGGATTGGCCTTATCATGCCAGTCTTGCCAACGTTATTACGTGAATTTATTGCTTCGGAAGATATCGCTAACCACTTTGGCGTATTGCTTGCACTTTATGCGTTAATGCAGGTTATCTTTGCTCCTTGGCTTGGAAAAATGTCTGACCGATTTGGTCGGCGCCCAGTGCTGTTGTTGTCATTAATAGGCGCATCGCTGGATTACTTATTGCTGGCTTTTTCAAGTGCGCTTTGGATGCTGTATTTAGGCCGTTTGCTTTCAGGGATCACAGGAGCTACTGGGGCTGTCGCGGCATCGGTCATTGCCGATACCACCTCAGCTTC...
Embodiment 2
[0131] Embodiment 2, DNA fragment preparation
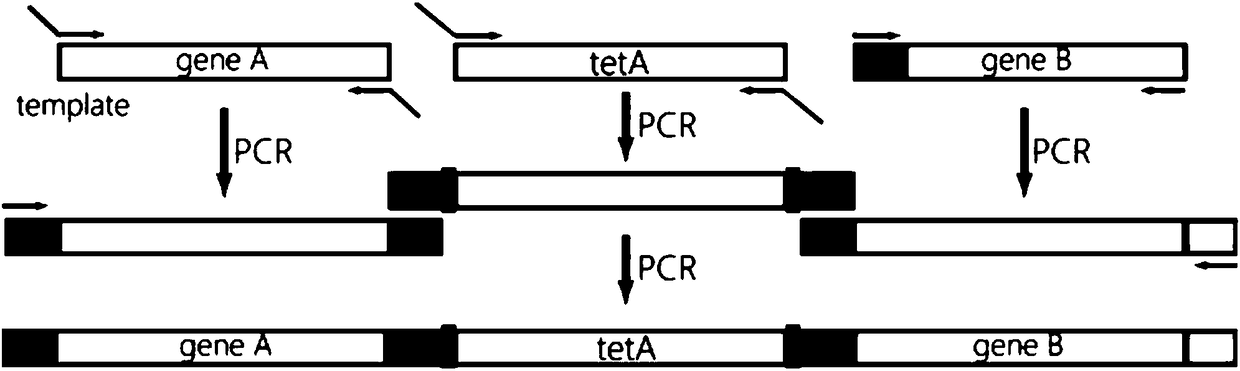
[0132] according to figure 2 As shown, first perform PCR to amplify the desired DNA sequence. PCR was divided into two rounds.
[0133] 1. The first round
[0134] (1), obtaining the fusion sequence of the crtE sequence and its upstream homologous sequence
[0135] Using the E.coli chromosome as a template, use primers crtE-H-F1 and crtE-H-R1 to amplify the upstream homologous sequence of crtE;
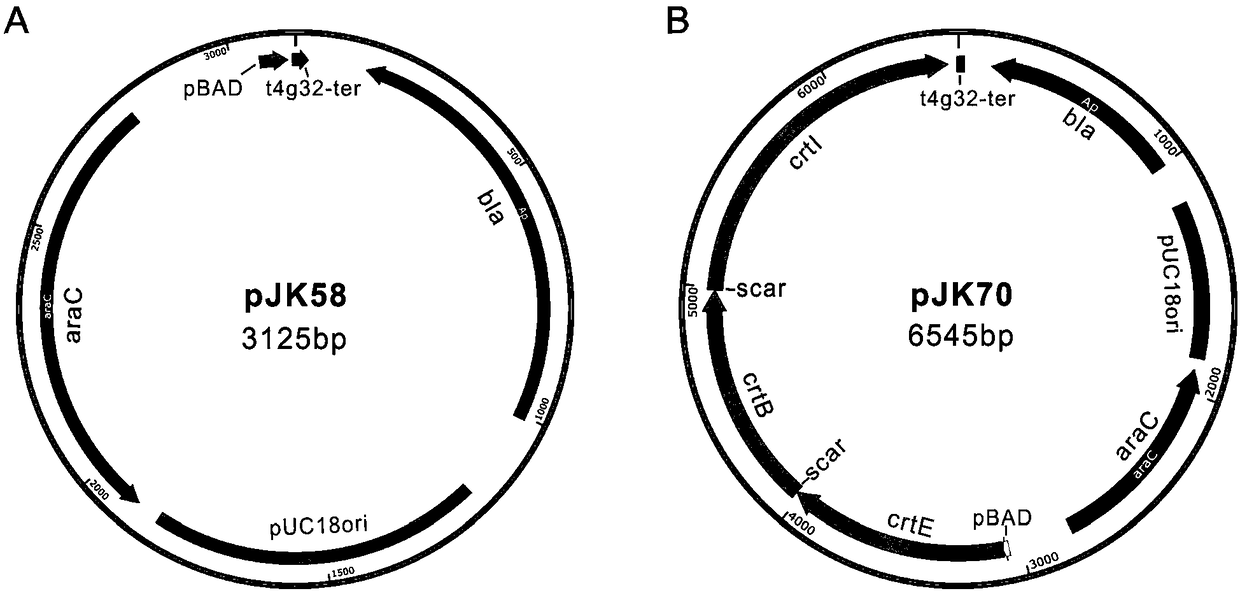
[0136] Using pJK70 as a template, use crtE-F, crtE-R1, crtE-R2 to amplify the crtE sequence;
[0137] Then, the crtE sequence and its upstream homologous sequence were fused together by the method of overlap-PCR.
[0138] (2), obtain the tetA gene sequence
[0139] Using pUCtetA as a template, the tetA gene was amplified using tetA-F1 and tetA-R1.
[0140] (3), obtaining the fusion sequence of crtB and its downstream homologous sequence
[0141] Using pJK70 as a template, use crtB-F1 and crtB-R1 to amplify the crtB sequence;
...
Embodiment 3
[0154] Embodiment 3, induce further verification
[0155] Since the inventor used the assembly of crtE, crtB and crtI as an example, the expression of these three genes can produce lycopene, the inventor cloned the assembled sequence into the vector pJK58, transformed it into E. coli, and induced it with arabinose Express.
[0156] The result is as Image 6 As shown, it can be seen that lycopene is expressed.
[0157] Furthermore, the inventors lysed the bacteria, extracted them with ethyl acetate, and performed LC-MS characterization. The results showed that lycopene was produced. This shows that the inventor's assembling method is correct and can be popularized.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com