Tumor marker STAMP-EP5 based on methylation modification
A methylation and tumor technology, applied in the direction of recombinant DNA technology, biochemical equipment and methods, microbial measurement/testing, etc., can solve problems such as misdiagnosis, insufficient sensitivity and specificity, difficult to use standards, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Embodiment 1, for the nucleic acid sequence detected by STAMP-EP5
[0073] The sequence of the STAMP-EP5 tumor marker is provided, as shown in the following SEQ ID NO: 1 (chr22: 46658718-46659169 / hg19), where the underline indicates that the base is a methylated CpG site, and the number below the underline indicates the position of the site serial number.
[0074]
[0075] The above sequence of SEQ ID NO:1 is treated with bisulfite and the sequence is as follows: SEQ ID NO:2 (wherein Y represents C or U):
[0076]
[0077]
[0078] The reverse complementary sequence of the nucleotide sequence shown in the above SEQ ID NO:1 is as follows: SEQ ID NO:3:
[0079] CG T CG GC CG CCTCC CG CTGC CG C CG GTTCCT CGCG GGGCACAAGC CG CCGTC
[0080] TC CG GGG CG CC CG GTGGCCTCCTCAGGGG CG CCC CG GGCCT CG GGGTG CGCG
[0081] G CG GC CGCG CCCTCCTCAGTCTG CG GCCCAG CGCG GTG CG GG CG GG CG G CG C
[0082] TGTCCTGAG CG GC CGCG GC...
Embodiment 2
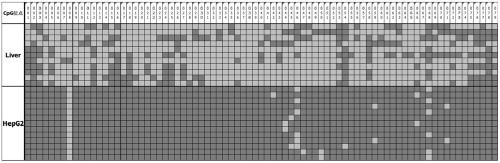
[0098] Example 2. Differences in the methylation of STAMP-EP5CpG sites between tumor cells and non-tumor cells——bisulfite-treated sequencing method (BSP-Bisulfite Sequencing PCR)
[0099] 1. Extract the genomic DNA of the liver cancer cell line HepG2 and the normal liver cell line;
[0100]2. Treat the extracted HepG2 and normal liver cell line genomic DNA with bisulfite, respectively, as templates for subsequent PCR amplification;
[0101] 3. Design amplification primers according to the sequence of SEQ ID NO: 1, design primers (SEQ ID NOs: 5-6; Table 1), and perform amplification by conventional methods.
[0102] 4. After PCR amplification, 2% agarose gel electrophoresis was used to detect the specificity of the PCR fragment, the gel was cut to recover the target fragment, ligated and inserted into the T vector, transformed into competent E. 10 clones were picked for Sanger sequencing.
[0103] Table 1, BSP primers
[0104]
[0105] BSP verification of the methylation ...
Embodiment 3
[0106] Example 3. Differences in the methylation of STAMP-EP5 CpG sites between tumor cells and non-tumor cells—pyrosequencing
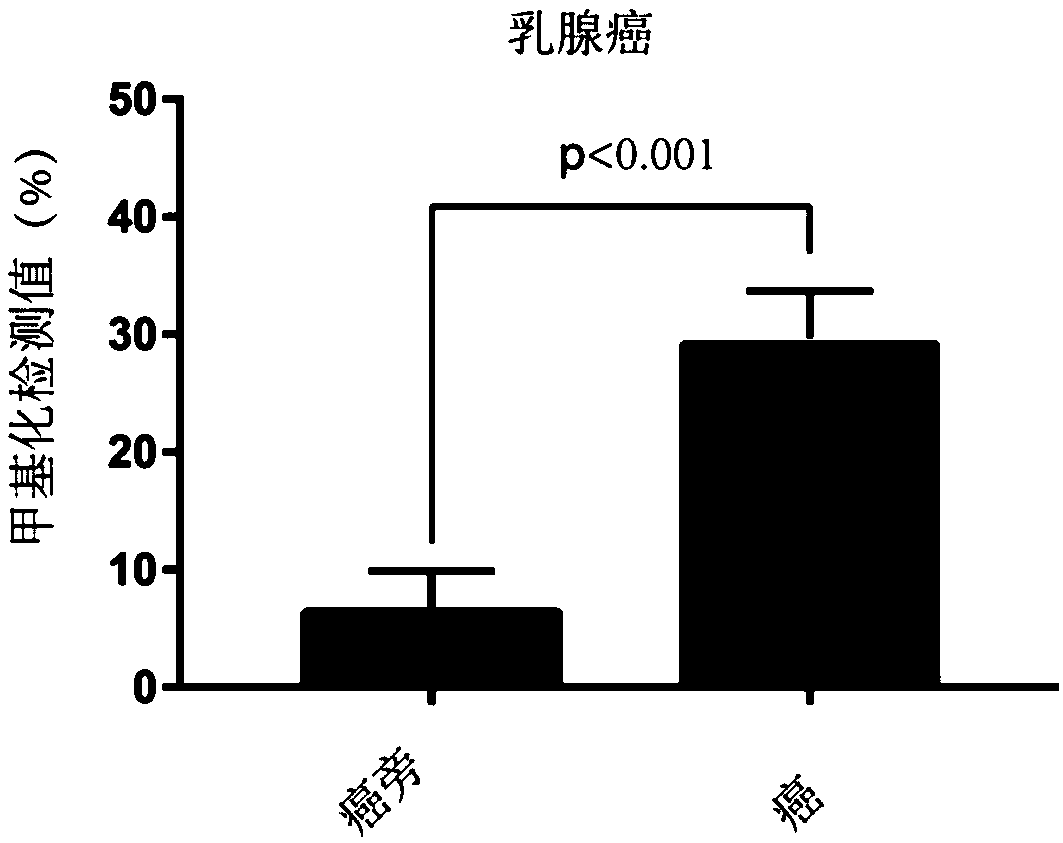
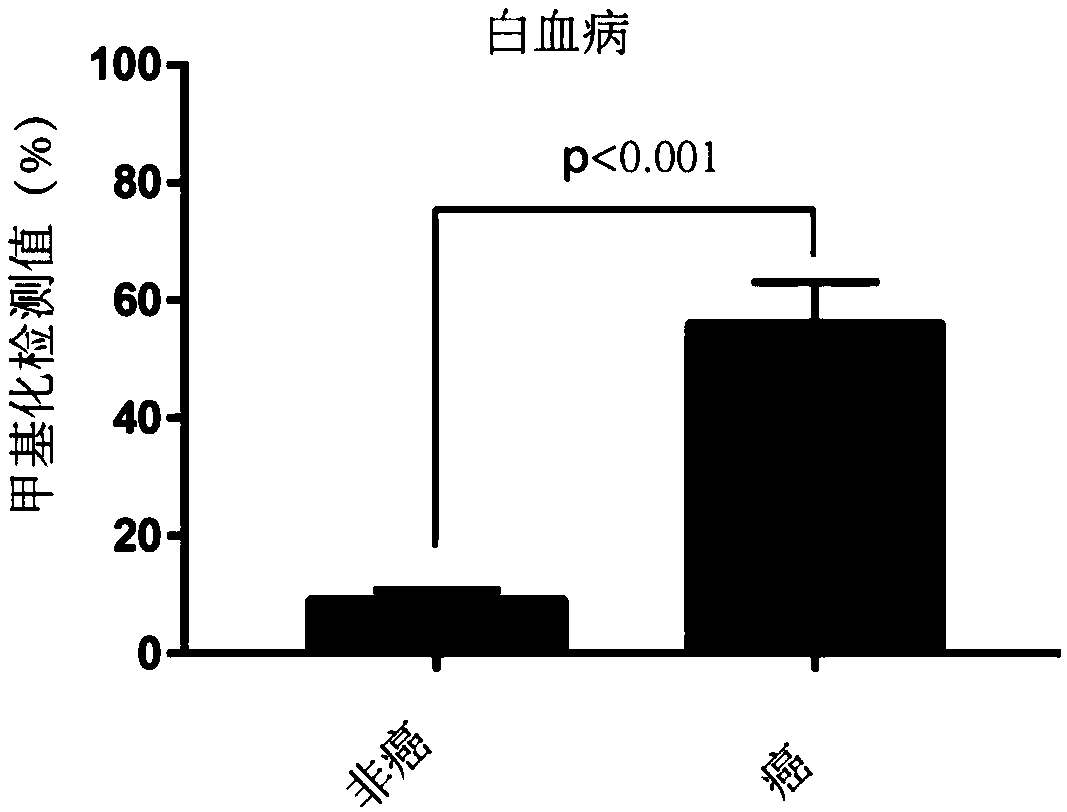
[0107] 1. Obtain clinical samples: Obtain para-cancer / non-cancer-cancer tissue samples from the clinic, para-cancer / non-cancer samples are used as the control group, and cancer tissue samples are used as the tumor detection experimental group;
[0108] 2. DNA extraction: extract the DNA of the experimental group and the control group respectively; this experiment uses the phenol-chloroform extraction method, but is not limited to this method;
[0109] 3. Bisulfite treatment: treat the extracted DNA samples with bisulfite, and operate in strict accordance with the steps; in this experiment, EZ DNA Methylation-Gold Kit from ZYMO Research Company, Cat. No. D5006 was used, but not limited to this kit;
[0110] 4. Primer design: According to the characteristics of STAMP-EP3 sequence SEQ ID NO: 1, design PCR amplification primers and pyrosequencing primers...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com