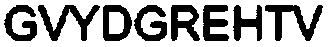T cell receptors
A cell receptor, surface plasmon technology, applied in animal cells, vertebrate cells, genetically modified cells, etc., can solve problems such as unknown functions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0098] Example 1 - Cloning of reference MAGE-A4 TCR α and β chain variable region sequences into pGMT7-based expression plasmids
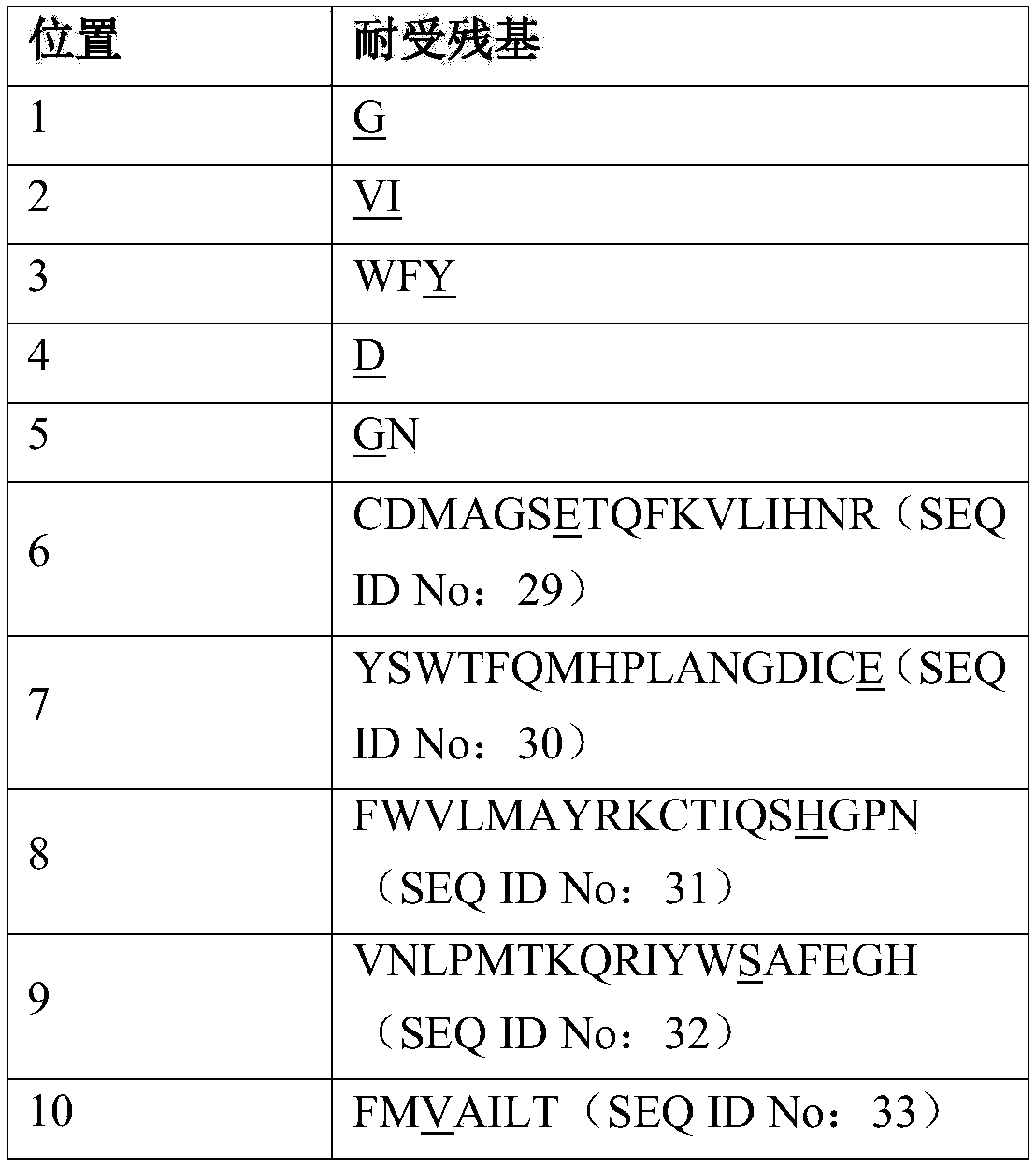
[0099] The parental MAGE-A4 TCR variable alpha and TCR variable beta domains of SEQ ID NO: 3 and SEQ ID NO: 4 were synthesized by standard methods described in (Molecular Cloning a Laboratory Manual 3rd Edition, Sambrook and Russell) Cloning into pGMT7-based expression plasmids containing Cα or Cβ. Plasmids were sequenced using an Applied Biosystems 3730xl DNA Analyzer. In the same manner, the reference MAGE-A4 TCR variable alpha and TCR variable beta domains of SEQ ID NO: 4 and SEQ ID NO: 5 were cloned.
[0100] The DNA sequence encoding the variable region of the TCRα chain was ligated into pEX956 cut with restriction enzymes. The DNA sequence encoding the variable region of the TCR beta chain was ligated into pEXb21, which was also cut with restriction enzymes.
[0101] The ligated plasmids were transformed into competent E. coli strain XL1-b...
Embodiment 2
[0102] Example 2 - Expression, refolding and purification of a soluble reference MAGE-A4 TCR
[0103] The expression plasmids containing the reference TCR α-chain and β-chain prepared in Example 1 were respectively transformed into Escherichia coli strain BL21pLysS, and a single ampicillin-resistant clone was incubated at 37°C in TYP (ampicillin 100 μg / ml ) medium grown to OD 600 was about 0.6-0.8, and then 0.5 mM IPTG was used to induce protein expression. Cells were harvested 3 hours after induction by centrifugation at 4000 rpm for 30 minutes in a Beckman J-6B. in MgCl 2 The cell pellet was lysed with 25 ml BugBuster (NovaGen) in the presence of DNaseI. Inclusion body pellets were recovered by centrifugation in a Beckman J2-21 centrifuge at 13000 rpm for 30 minutes. Three detergent washes were then performed to remove cellular debris and membrane components. Each inclusion body pellet was homogenised in Triton buffer (50 mM Tris-HCl pH 8.0, 0.5% Triton-X100, 200 mM NaC...
Embodiment 3
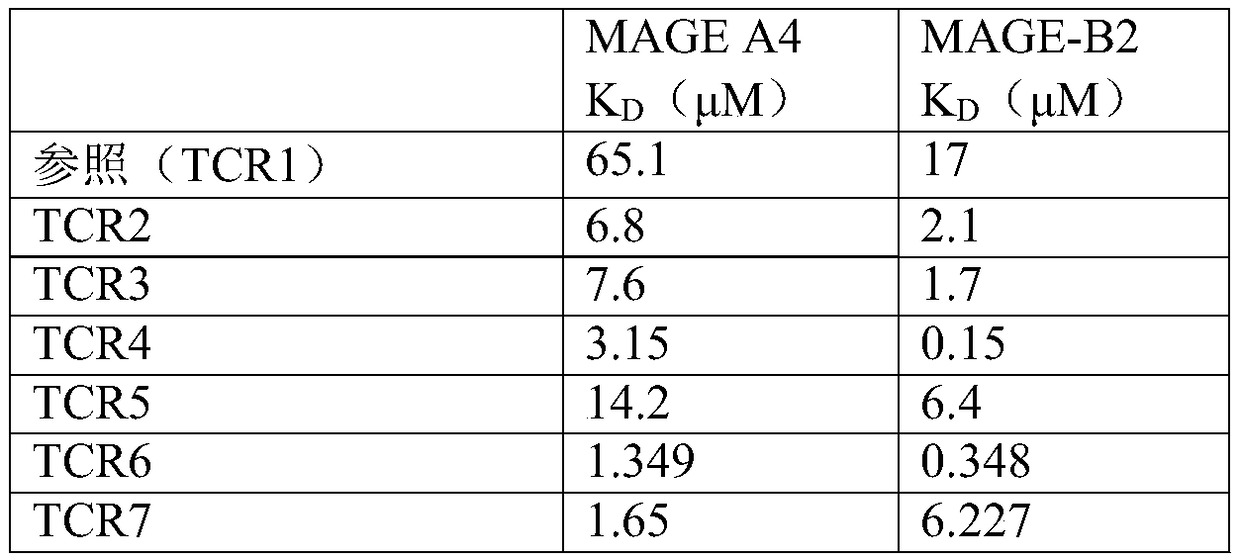
[0106] Example 3 - Binding Characterization
[0107] BIAcore analysis
[0108] Surface plasmon resonance biosensor (BIAcore 3000 TM ) can be used to analyze the binding of soluble TCRs to their peptide-MHC ligands. This is facilitated by the generation of soluble biotinylated peptide-HLA ("pHLA") complexes that can be immobilized to streptavidin-coated binding surfaces (sensor chips). The sensor chip contains four individual flow cells that are capable of simultaneously measuring the binding of T cell receptors to four different pHLA complexes. Manual injection of pHLA complexes allows easy manipulation of precise levels of immobilized class I molecules.
[0109] In vitro refolding of biotinylated class I HLA-A*0201 molecules from bacterially expressed inclusion bodies containing constitutive subunit proteins and synthetic peptides, followed by purification and in vitro enzymatic biotinylation (O'Callaghan et al. (1999) Anal. Biochem. 266:9-15). The HLA-A*0201-heavy chain...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com