Arrays for single molecule detection and uses thereof
An array and sequence technology, applied in the field of manufacturing and using spatially addressable low-density or high-density molecular arrays, which can solve the problems of complex samples causing difficulty, separation, and difficulty in resolving heterogeneous events.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0369] The present invention involves combining the preparation of single molecule arrays with performing assays on the single molecule arrays. Especially when either or both are combined with detection / imaging of individual probes on a substrate as described herein and assays based on counting single molecules or recording and measuring signals from single molecules.
[0370] The invention also provides software and algorithms for processing data from the methods described above.
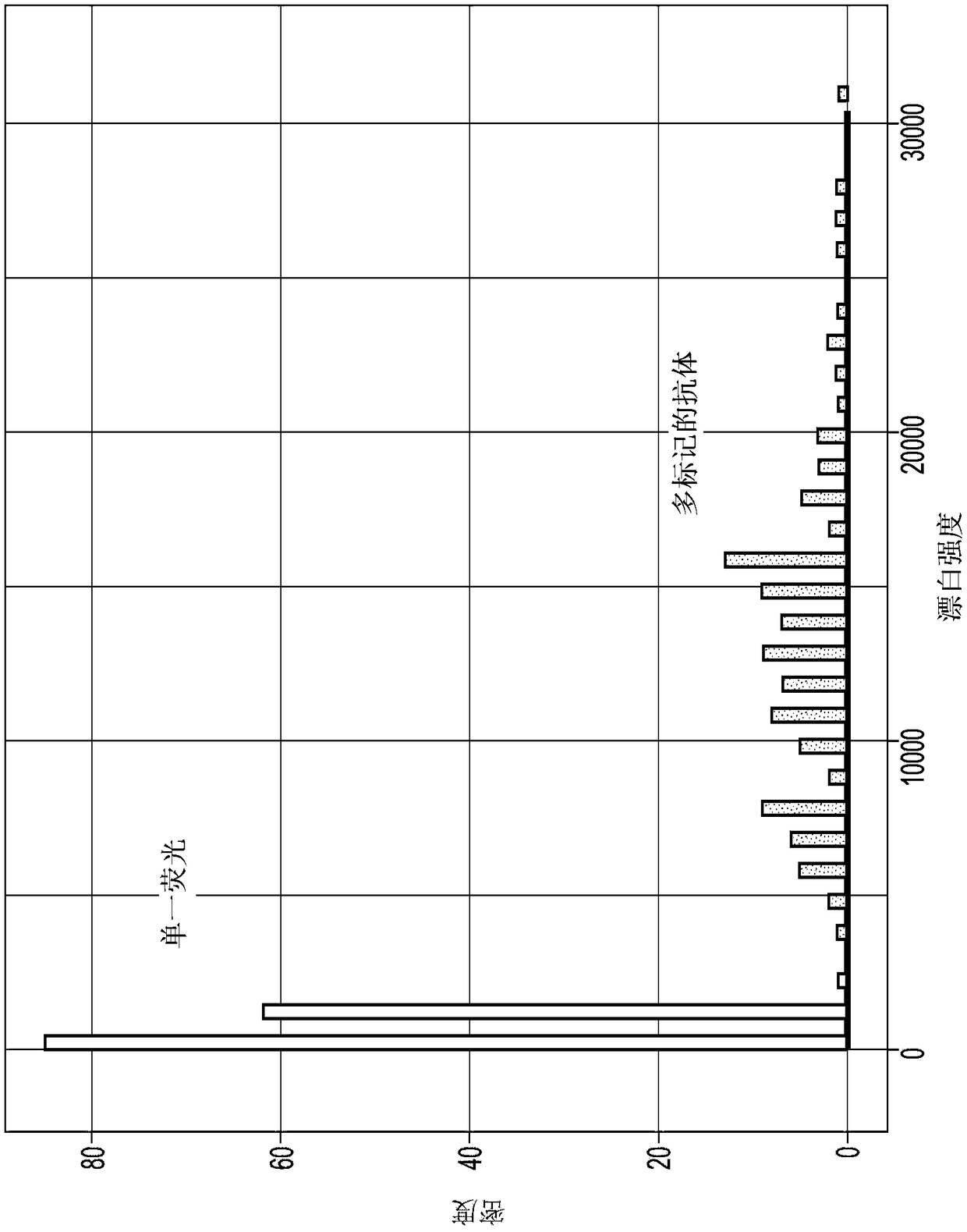
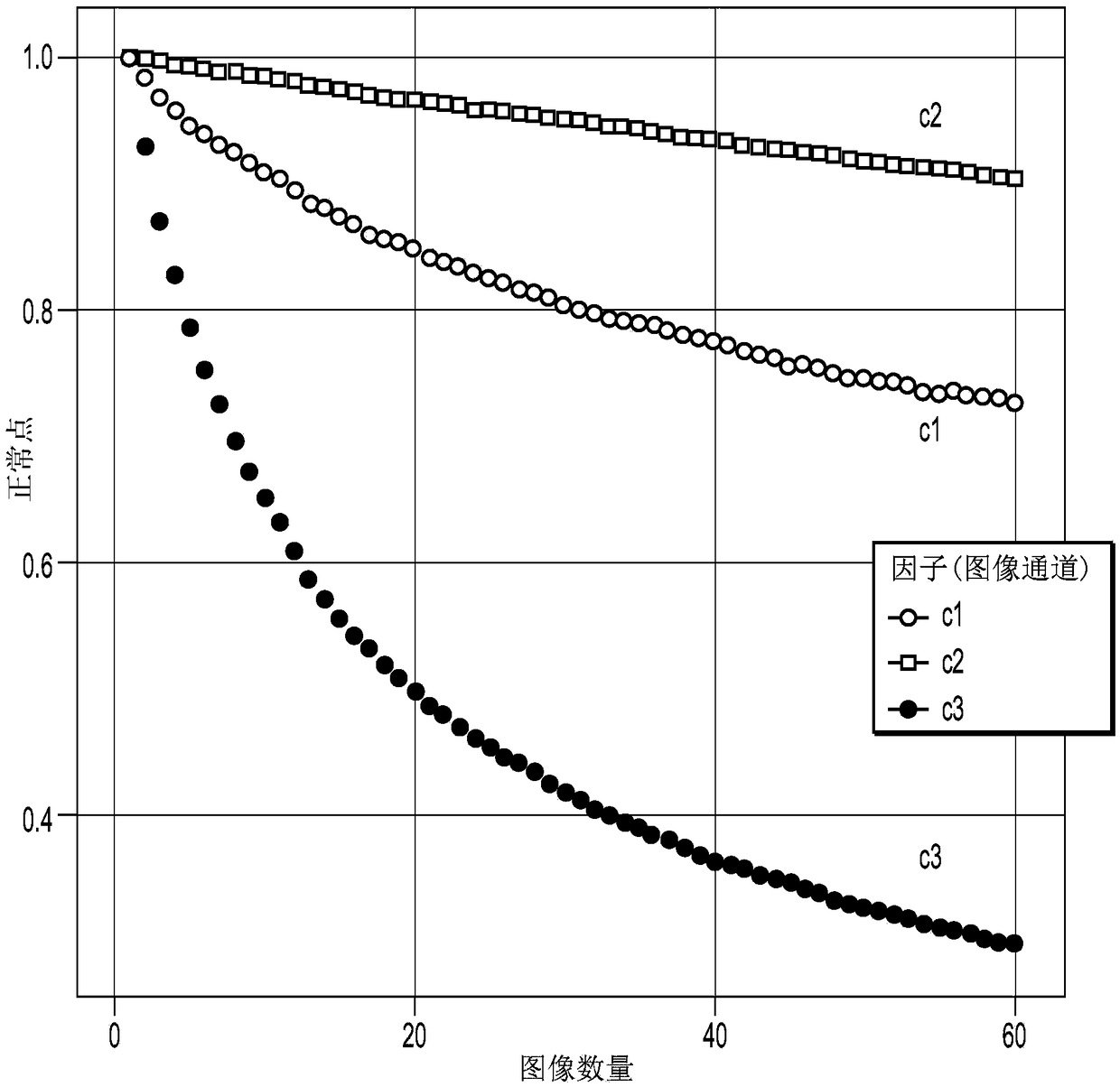
[0371] A system for detecting genetic variation according to the methods described herein includes a variety of elements. Some elements include converting a raw biological sample into a useful analyte. The analyte is then detected and data generated which is later processed into a report. Figure 19 Various modules that can be included in the system are shown in . Figure 20 More details of the various methods used to analyze the data (eg including image processing) are shown in . Analysis can ...
Embodiment approach
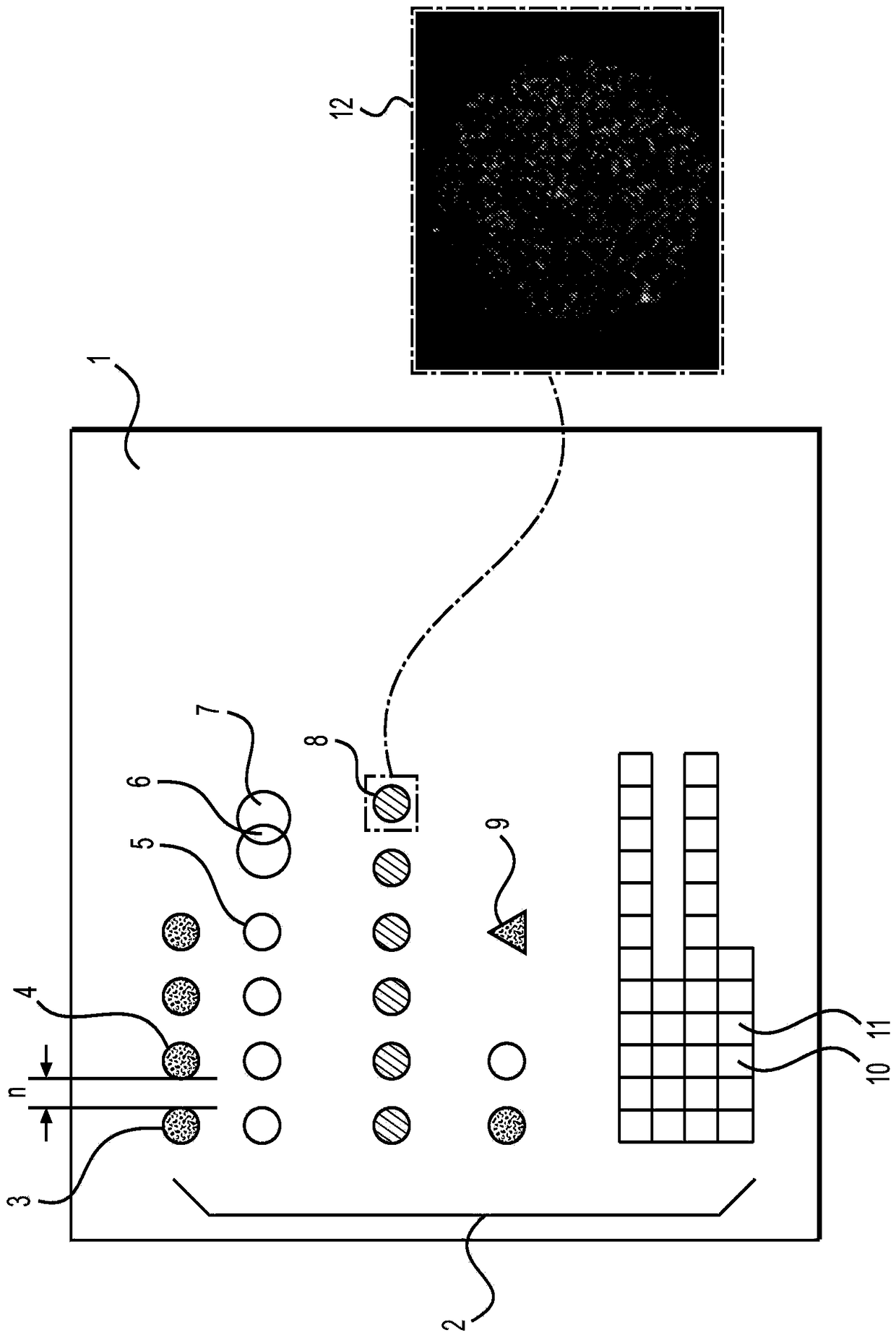
[0753] Figure 21 is an implementation of the assay used to quantify genomic copy number at two genomic loci. In this embodiment of the assay, 105 and 106 are target molecules. 105 contains the sequence corresponding to the first genomic locus "locus 1" corresponding to the query copy number (e.g., chromosome 21), and 106 contains the sequence corresponding to the second genomic locus "locus 2" corresponding to the query copy number (e.g., chromosome 18). sequence. Figure 21 An example of one probe set per genomic locus is included, but in some embodiments of the assay, multiple probe sets can be designed to interrogate multiple regions within a genomic locus. For example, greater than 10, greater than 100, or greater than 500 probe sets corresponding to chromosome 21 can be designed. Figure 21 Only a single probe set for each genomic locus is illustrated, but importantly the scope of the invention allows the use of multiple probe sets for each genomic locus. Figure 21 ...
Embodiment 1
[0911] Example 1 - Cleaning the Substrate
[0912] The following steps are preferably performed in a clean room. Thoroughly clean pure white glass plates / slides (Knittel Glazer, Germany) (which can be polished for flatness) or Spectrosil slides by, for example, ultrasonication in a surfactant solution (2% Micro-90) for 25 minutes. surface, washed in deionized water, rinsed thoroughly with milliQ water, and immersed in 6:4:1 milliQ H 2 O:30%NH 4 OH:30%H 2 o 2 or immersed in H 2 SO 4 / CrO 3 1.5 hours in the cleaning solution. After cleaning, the plates are rinsed and stored in a dust-free environment, for example under milliQ water. The top layer of the mica substrate was peeled off by covering with scotch tape and quickly tearing off the layer.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com