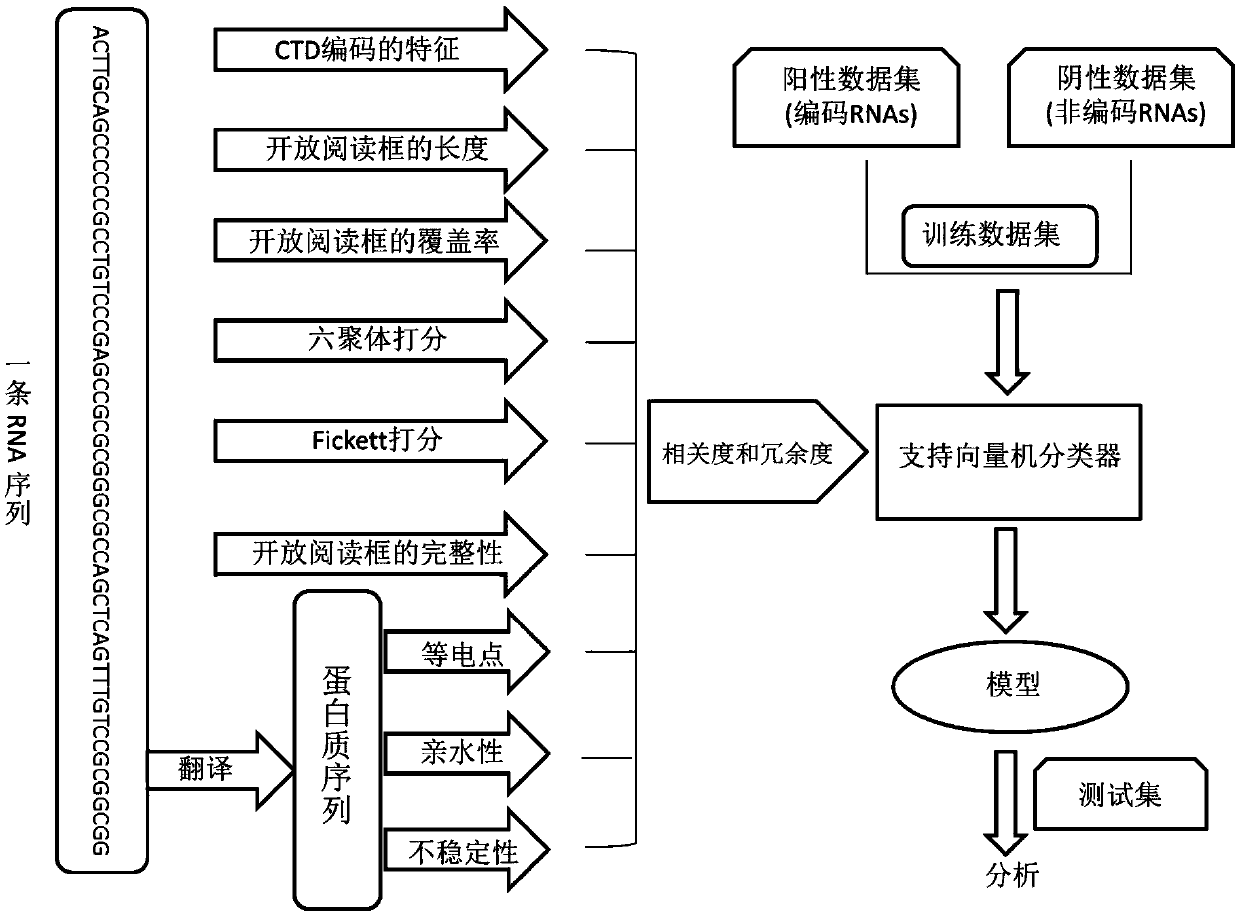
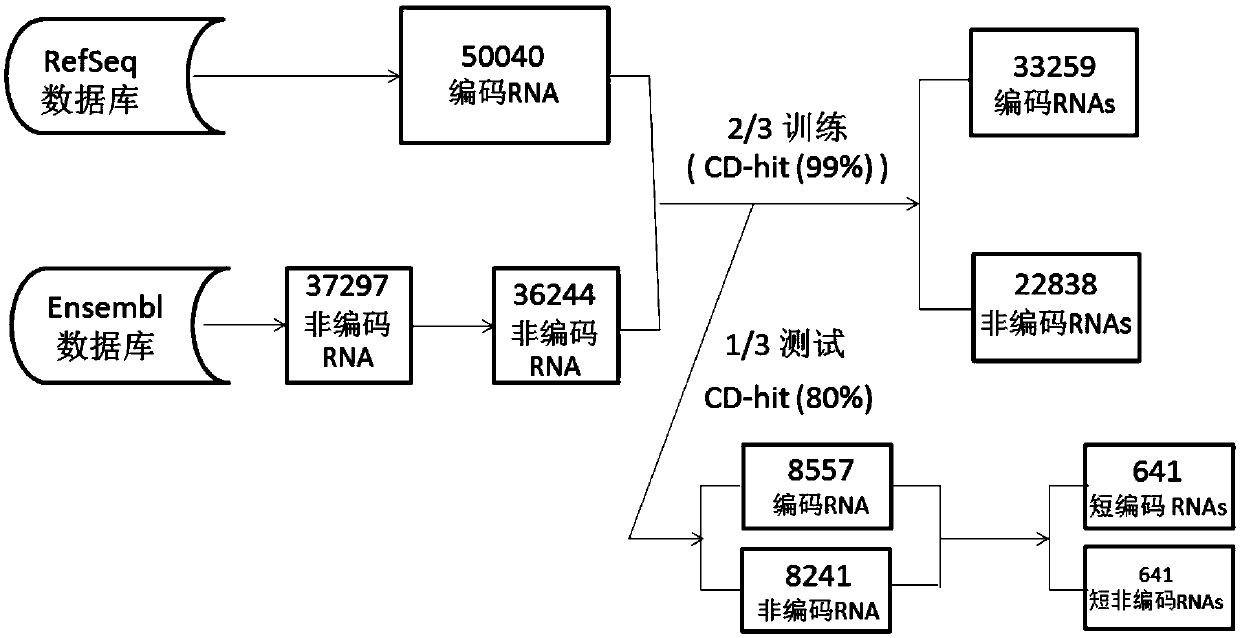
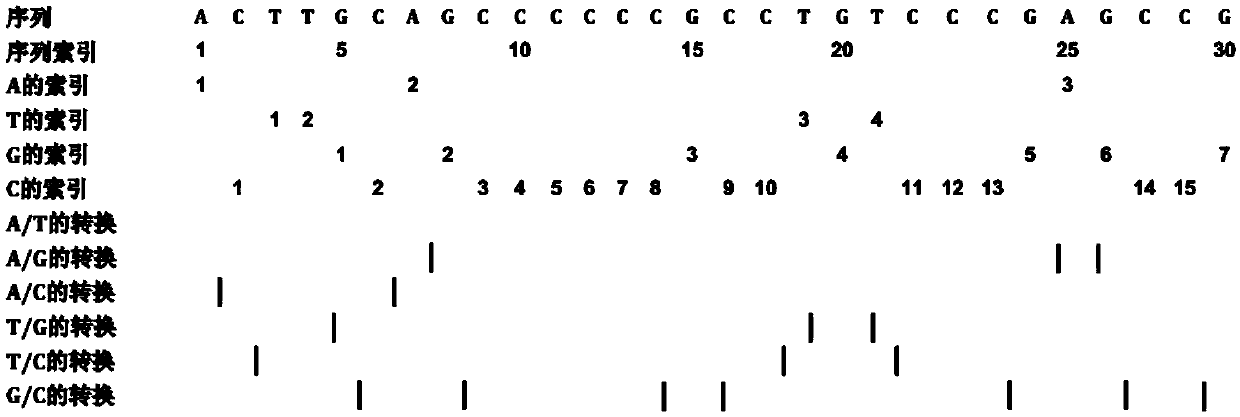
Method for predicting RNA coding potential
A prediction method and potential technology, applied in the field of prediction of RNA coding potential, can solve problems such as low prediction accuracy and fitting risk, achieve good species universality, high accuracy, and reduce species dependence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0058] The present invention uses CPPred to test the data of human, mouse, zebrafish and Saccharomyces cerevisiae, and compares it with the test results of existing CPAT, CPC2, PLEK and sORF finder tools.
[0059] On the human test set (including long and short sequences) and the human sORF test set, the prediction performance comparison results of different prediction tools are shown in Table 1 and Table 2. It can be seen from Table 1 and Table 2 that whether it is the human test set or the human sORF test set, CPPred is better than CPAT and CPC2, but slightly worse than PLEK. This is because of redundancy between PLEK's training set and human test set.
[0060] Table 1: Comparison of CPPred with CPAT, CPC2, PLEK humans on the test set
[0061]
[0062] Table 2: Comparison of CPPred with CPAT, CPC2, PLEK, sORF finder on the human sORF test set
[0063]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com