Escherichia coli for producting farnesene
A technology for producing farnesene and bacteria, which is applied in the field of genetic engineering and can solve problems such as low fermentation levels
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
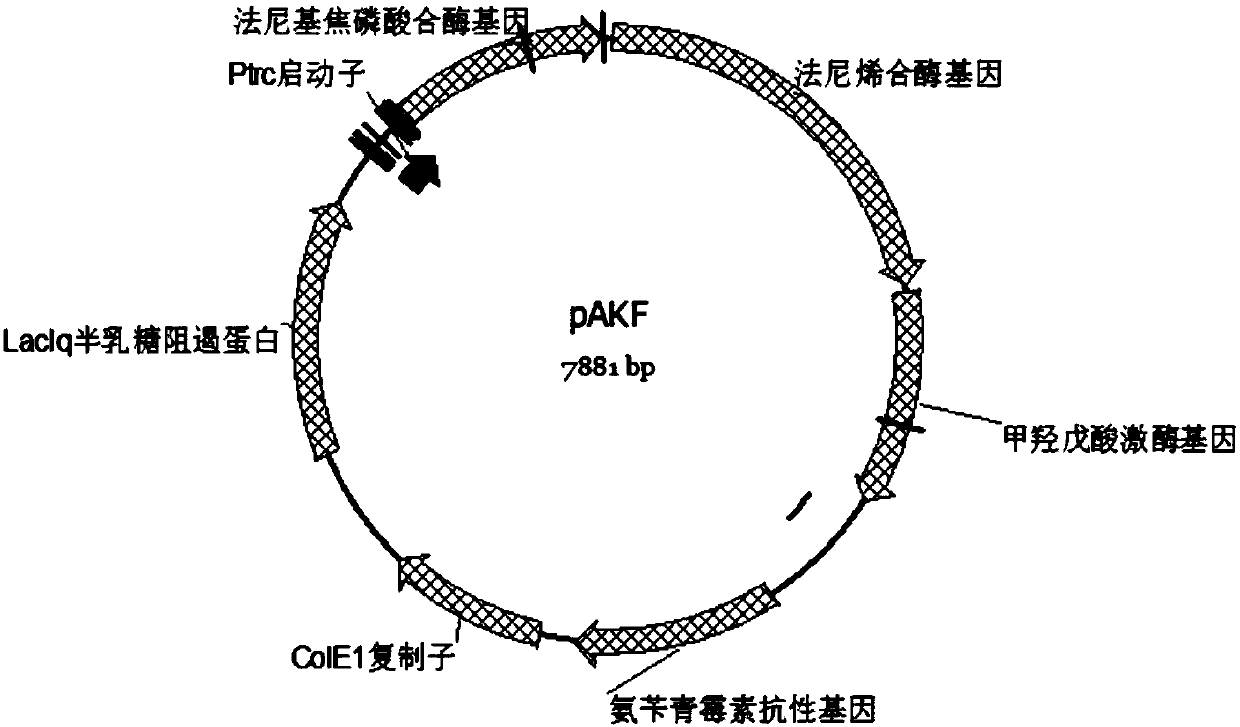
[0050] Embodiment 1: Construction of plasmid pAKF
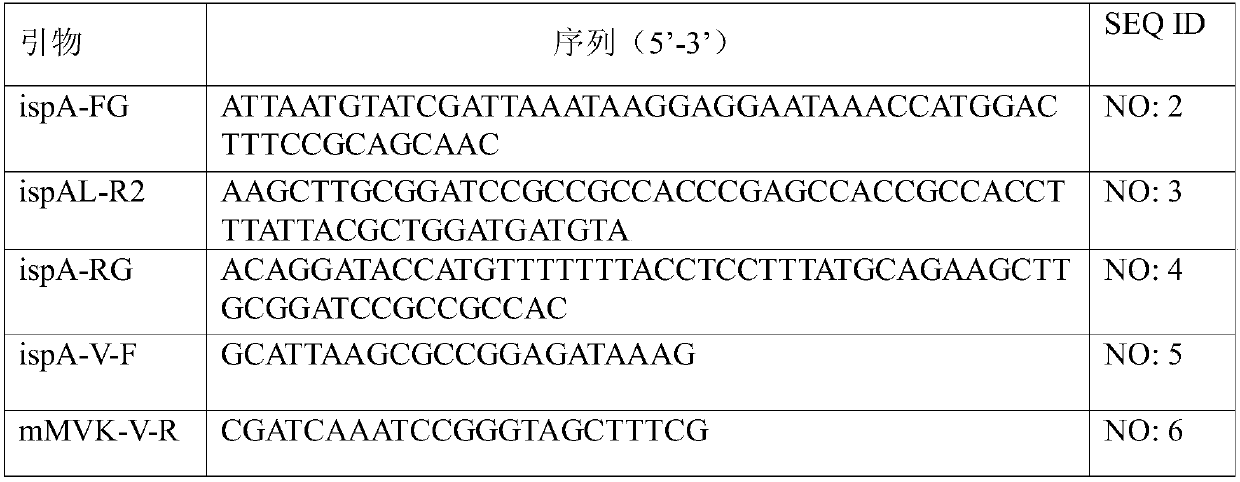
[0051] The primer sequence information used in the construction of plasmid pAKF is shown in Table 1.
[0052] Table 1. Primer sequences
[0053]
[0054] In Table 1, "-F" in the name stands for forward direction; "-R" stands for reverse direction.
[0055] 1.1 Using the Escherichia coli MG1655 genome as a template and ispA-FG / ispAL-R2 as primers, PCR amplifies the ispAL fragment, about 969 bp (amplification of homology arm + ispA + linker). PCR conditions: denaturation at 95°C for 5 min, denaturation at 95°C for 30 s, annealing at 58°C for 30 s, extension at 68°C for 1 min, 32 cycles, 10 min at 68°C, and 10 min at 16°C.
[0056] The ispAL fragment was recovered from the gel, and the ispAL fragment was used as a template and the ispA-FG / ispA-RG was used as a primer to amplify the ispALG fragment by PCR, which was about 1004bp (amplified homology arm+ispA+linker+homology arm). PCR conditions: denaturation at 95°C for 5 min,...
Embodiment 2
[0059] Embodiment 2: Construction of β-farnesene producing bacteria
[0060] 2.1 The plasmids PHGFH, PAGEs, and pAKF in Example 1 were directly co-transformed into Escherichia coli CIBTS1758 by electroporation, and the strain CIBTS1758 / PHGFH / PAGEs / pAKF was obtained, which was named CIBTS2509C.
[0061] 2.2 The plasmids PHGFH, PAGEs, and pAK in Example 1 were co-transformed into Escherichia coli CIBTS1758 by electroporation, and the strain CIBTS1758 / PHGFH / PAGEs / pAK was obtained as a control strain.
Embodiment 3
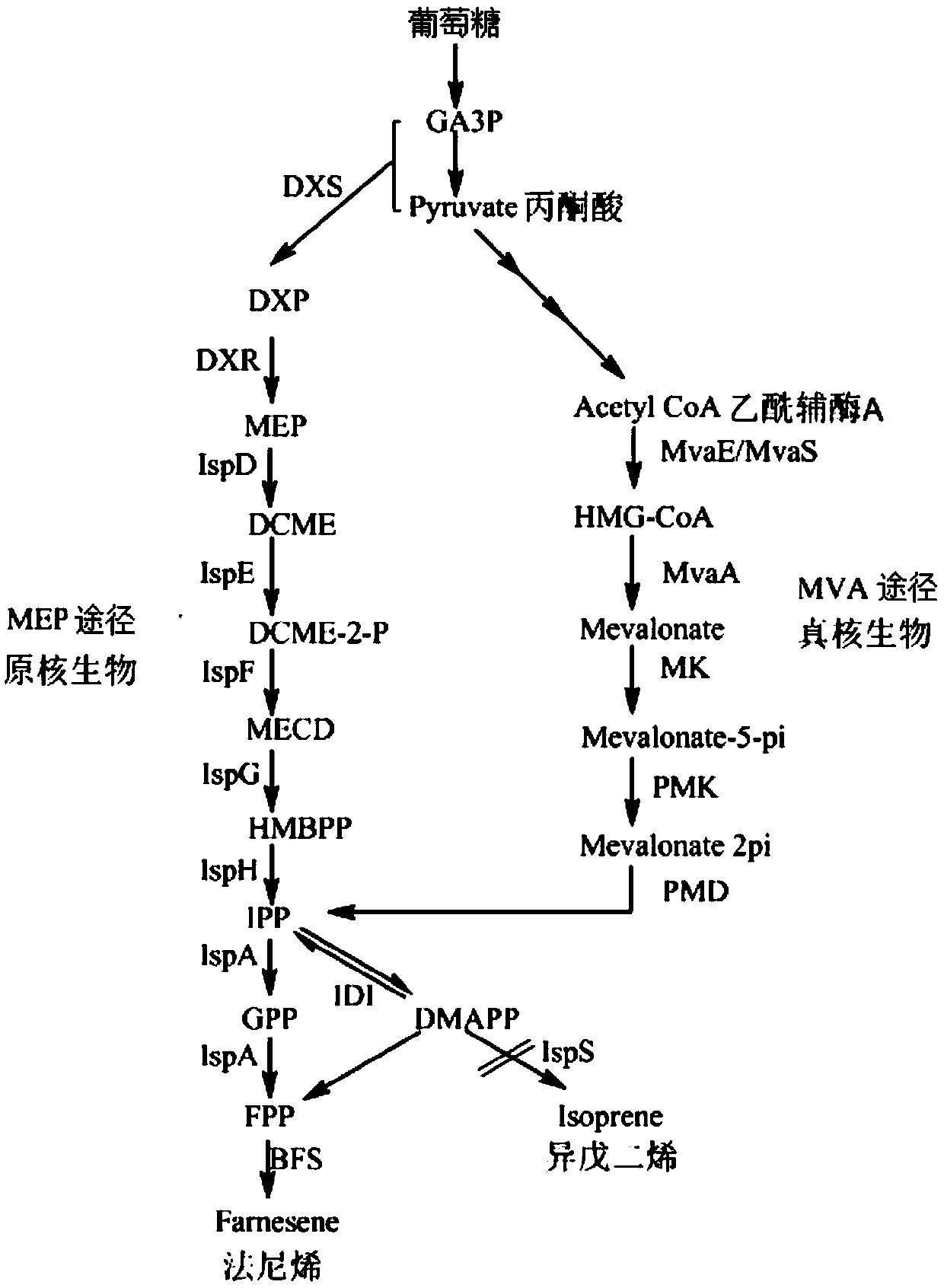
[0062] Example 3: Fermentative production of β-farnesene by bacterial strains
[0063] 3.1 Culture and fermentation of strain CIBTS2509C
[0064] Pick a single colony of CIBTS2509 into the LB medium containing 100 μg / ml ampicillin, 34 μg / ml chloramphenicol and 100 μg / ml spectinomycin, culture overnight at 37°C and 220 rpm; then inoculate at an inoculum of 8% v / v In the V7S medium containing the above three antibiotics, continue to culture at 37°C and 220rpm for 8-12h; then inoculate the V7E medium containing the above three antibiotics and 0.1mM IPTG according to the inoculum size of 8% v / v, at 37°C , 220rpm to continue culturing for 8-12h; add 1 / 5 volume of decane to continue culturing for 4h. Centrifuge at 12,000 rpm for 10 min, collect the decane phase, and send it to GC-MS for analysis.
[0065] The GC analysis method of farnesene is as follows: Agilent 7890A gas chromatograph equipped with 5975MS; chromatographic column Agilent HP-1 (30mx0.25mm, 0.25μm); injection volum...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com