Coronavirus resistant clone pig and preparation method thereof
A coronavirus and resistance technology, applied in the genetic field, can solve the problem of blocking a variety of coronavirus infections, and achieve the effect of resisting swine coronavirus infection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0021] A method for preparing a coronavirus-resistant cloned pig based on CRISPR / Cas9 gene editing technology, comprising the following steps:
[0022] (1) Obtain gRNA sequences specifically targeting exon 2 and exon 3 of the pAPN gene, the nucleotide sequences of which are shown in sequence as SEQ ID No.1-2;
[0023] Prepare the CRISPR / Cas9 targeting vector corresponding to the gRNA sequence. The preparation method of the CRISPR / Cas9 targeting vector is: digest the PX330 vector with the restriction endonuclease BbsI; synthesize the second exon and the third exon respectively The gRNA positive strand and the complementary strand DNA of the exon, with sequences matching the sticky ends formed by the digested PX330 vector at both ends; the gRNA positive strands of the 2nd exon and 3rd exon with the The complementary strand DNA is mixed separately, and then the mixed sequence is placed in water with a temperature of at least 98-100°C and boiled for 2.5-3.5min, then allowed to sta...
Embodiment 1
[0038] Construction of pAPN gene knockout vector
[0039] 1. Selection and synthesis of targeting sites.
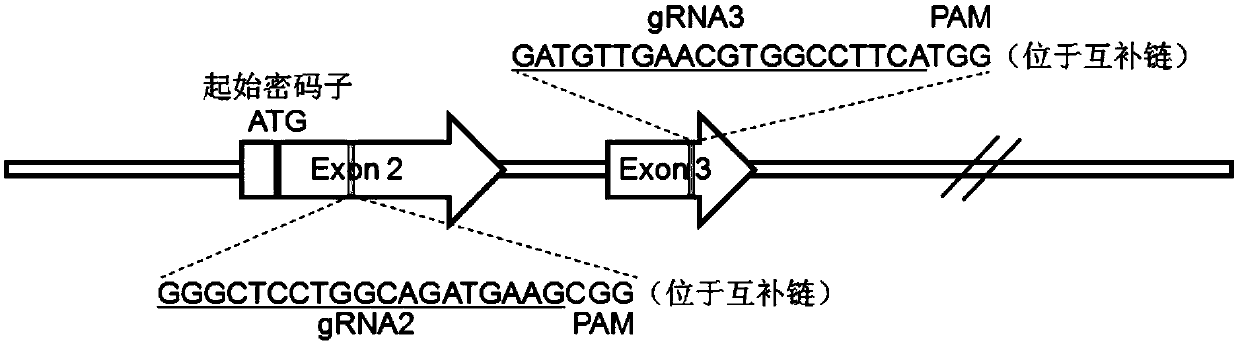
[0040] Select the CRISPR targeting site according to the sequence rules of GN(18-20)GG. The site must be located on the exon downstream of ATG and located in the first 1 / 3 of the coding region, so as to ensure that the introduction of mutations can lead to a more complete protein inactivation. The two selected targeting sites are located on exon 2 and exon 3, respectively, named gRNA2 and gRNA3.
[0041] The lengths are 19bp and 20bp respectively, and the sequences are GGGCTCCTGGCAGATGAAG and GATGTTGAACGTGGCCTTCA respectively. The structure of pAPN gene and the selected target site information are as follows: figure 1 shown. The oligonucleotides used to express gRNA were synthesized according to the selected targeting sites, and the sequences are shown in Table 1.
[0042] Table 1
[0043]
[0044] 2. Construction of pAPN targeting vector.
[0045] The PX330 vec...
Embodiment 2
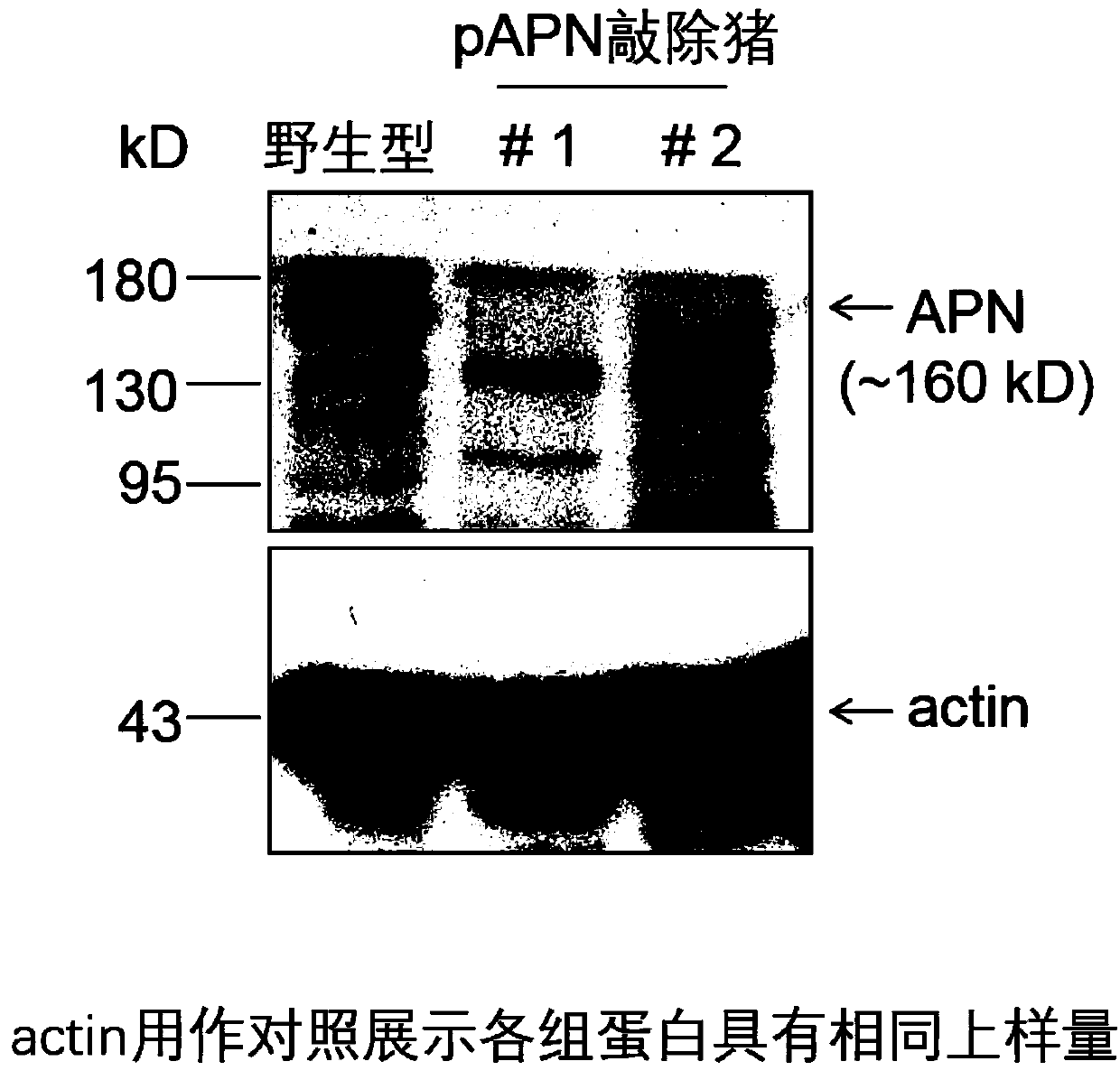
[0047] Example 2: Screening of pAPN knockout cells
[0048] 1. The primary isolated 30-day pig fetal fibroblasts were cultured to more than 80% confluence, digested and centrifuged to remove the cell culture medium, and then resuspended the cells in PBS to adjust the cell concentration to 10 5 each / 100μl PBS. Take 100 μl of cell suspension, mix with 10 μg of targeting plasmid, and place in Lonza Nucleofector for electroporation and transfection. The electric shock program was selected as A330. After electroporation, the cells were divided into ten 10cm culture dishes, with about 1000 cells per dish, and the culture medium was DMEM+15%FBS at 37°C, 5%CO 2 cultured in an incubator.
[0049] 2. After 10 days of transfection, the cells form a single clone of appropriate size. At this time, single cell clones need to be picked and transferred to a 48-well plate. Continue to culture to 24 wells.
[0050] 3. Remove 1 / 10 of the cells by centrifugation to remove the culture medium,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com