Method for increasing editing efficiency of base editing system
A base editing and editing technology, applied in botany equipment and methods, biochemical equipment and methods, genetic engineering, etc., can solve the problems of low efficiency and limitations of HDR
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
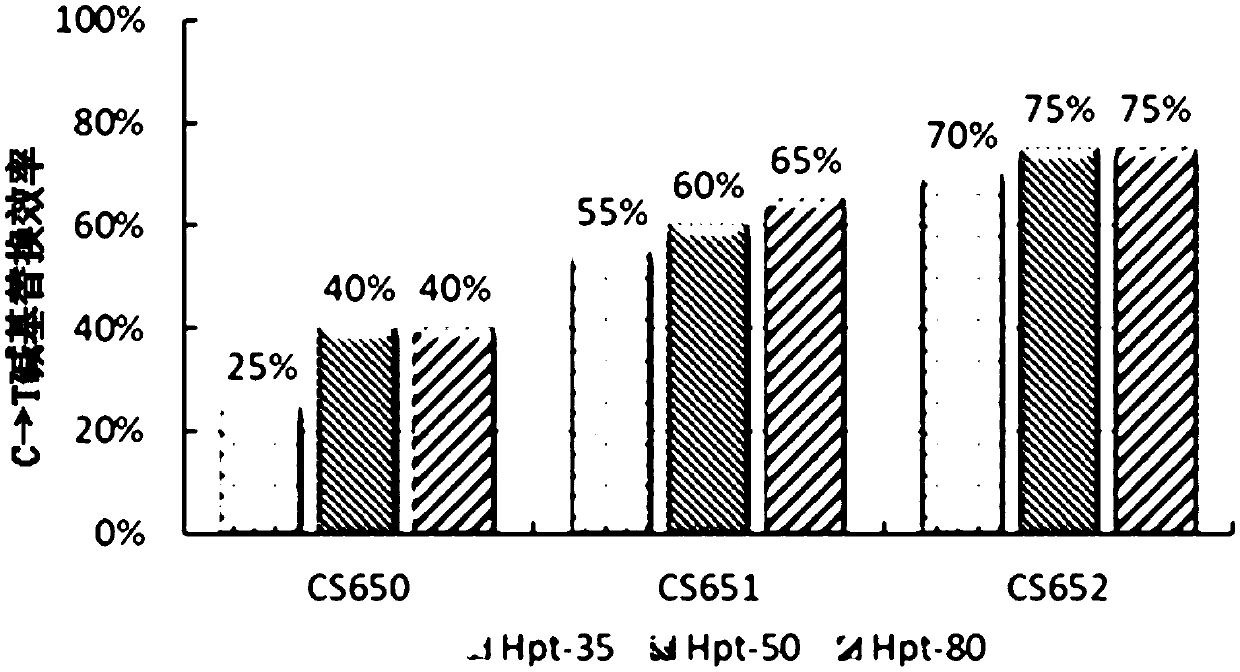
[0042] Example 1. High-concentration hygromycin screening improves the C→T base substitution efficiency of the target in the SpCas9n(D10A)&PmCDA1&UGI base editing system
[0043] 1. Construction of gene editing vector
[0044] The inventors of the present invention constructed the vector SpCas9n&PmCDA1&UGI.
[0045] The nucleotide sequence of the vector SpCas9n&PmCDA1&UGI (circular) is shown in sequence 1 in the sequence listing. In sequence 1, from 5' to 3', positions 131 to 467 are the nucleotide sequence of the OsU3 promoter, positions 474 to 550, positions 647 to 723 and positions 820 to 896 are all pre-tRNA Nucleotide sequence, the 551st to 570th is the nucleotide sequence of CS650, the 571st to 646th, 744th to 819th and 917th to 992th are the nucleotide sequence of the sgRNA backbone, the 724th to 743rd It is the nucleotide sequence of CS651, the 897th to 916th is the nucleotide sequence of CS652, the 993rd to 1283rd is the nucleotide sequence of the OsU3 terminator, a...
Embodiment 2
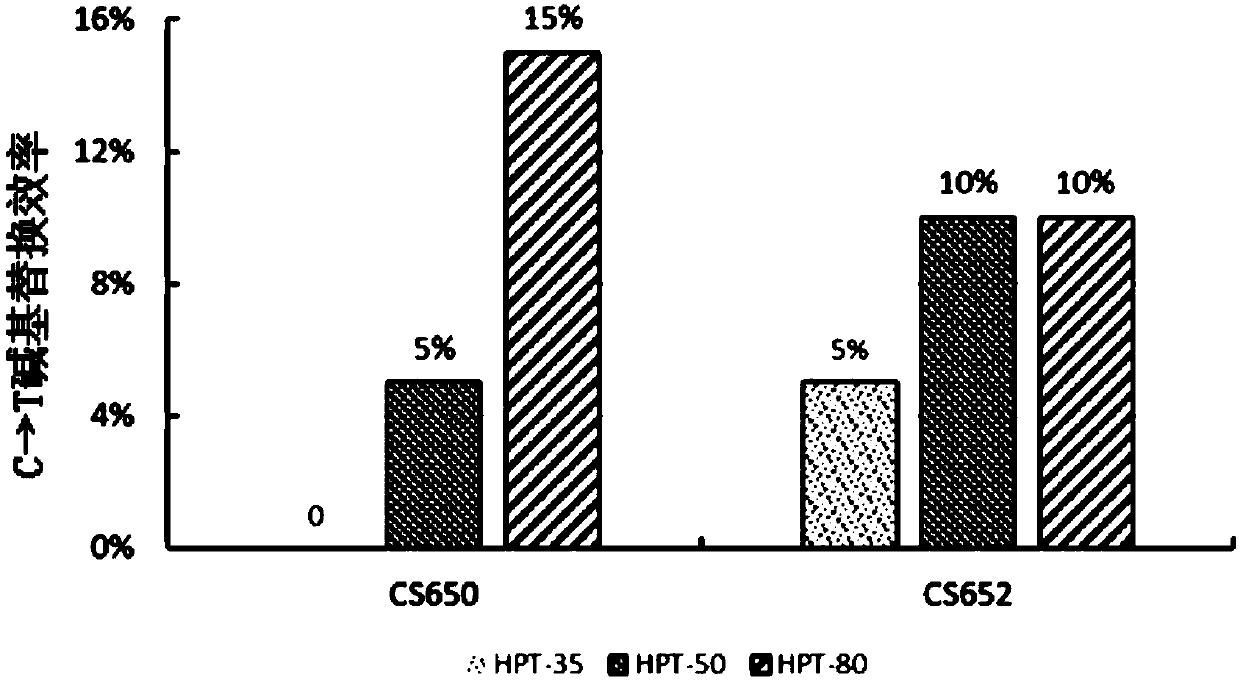
[0063] Example 2. High-concentration hygromycin screening improves the C→T base substitution efficiency of the target site by the rAPOBEC1&SpCas9n&UGI base editing system
[0064] 1. Construction of gene editing vector
[0065] The inventors of the present invention constructed the vector rAPOBEC1&SpCas9n&UGI.
[0066] The nucleotide sequence of the vector rAPOBEC1&SpCas9n&UGI (circular) is shown in sequence 2 in the sequence listing. In sequence 2, from 5' to 3', the 131st to 467th is the nucleotide sequence of the OsU3 promoter, the 474th to 550th and the 820th to 896th are all the nucleotide sequences of pre-tRNA, and the 1st Positions 551 to 570 are the nucleotide sequence of CS650, positions 571 to 646 and positions 917 to 992 are the nucleotide sequences of the sgRNA backbone, positions 897 to 916 are the nucleotide sequence of CS652, positions 993 to 1283 The position is the nucleotide sequence of the OsU3 terminator, the 1290th to 3003rd is the nucleotide sequence of...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com