System and method for genome editing
A genome editing and genome technology, applied in the field of genetic engineering, can solve problems such as genome editing of the AacC2c1 system that has not been proven
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0148]Example 1. In vitro analysis of AaC2c1 nuclease activity.
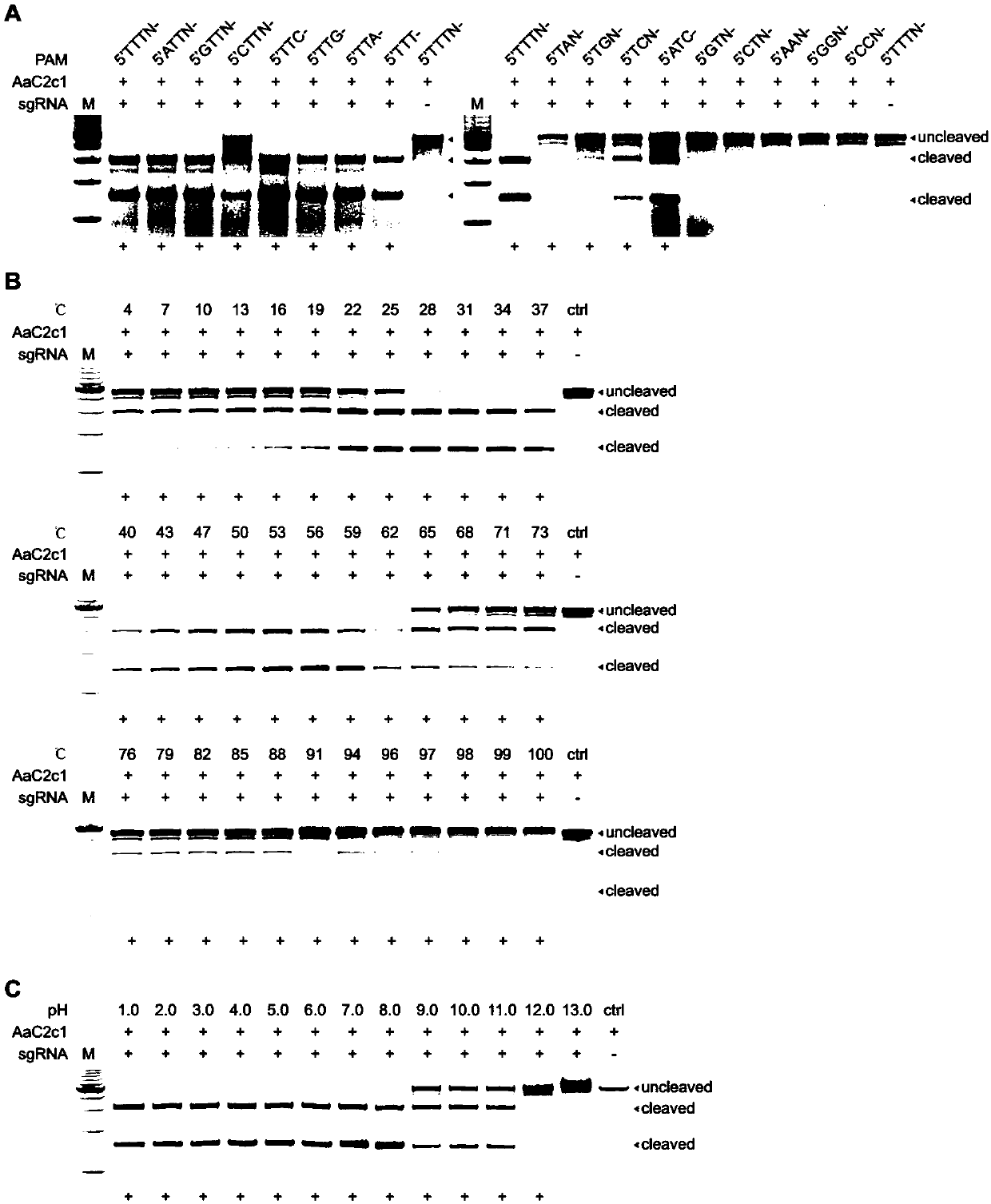
[0149] First, the PAM sequence of the present invention from A. acidiphilus C2c1 was identified by in vitro nucleic acid cleavage. figure 1 A shows cleavage of AaC2c1 and sgRNAs targeting loci with various PAMs. The symbol "+" below the graph indicates strong in vitro cleavage activity. The results show that the PAM of AaC2c1 can be 5'TTTN-, 5'ATTN-, 5'GTTN-, 5'CTTN-, 5'TTC-, 5'TTG-, 5'TTA-, 5'TTT-, 5'TAN -, 5'TGN-, 5'TCN-, 5'ATC-.
[0150] Second, the temperature and acid-base tolerance of AaC2c1 were tested. figure 1 B shows the cleavage activity of AaC2c1 over a wide temperature range (4°C-100°C). figure 1 C Analysis of AaC2c1 cleavage activity at a wide range of pH values (pH1.0-pH13.0). The symbol "+" below the graph indicates strong in vitro cleavage activity. The results show that AaC2c1 can work at 4°C-100°C, and the cutting efficiency is higher at about 30°C-60°C. AaC2c1 can work at pH1.0-pH12....
Embodiment 2Aa
[0158] Example 2 Genome Editing Activity of AaC2c1 in Mammalian Cells
[0159] In this example, the genome editing activity of AaC2c1 in mammalian cells was detected. The target sequences used are shown in Table 3 below.
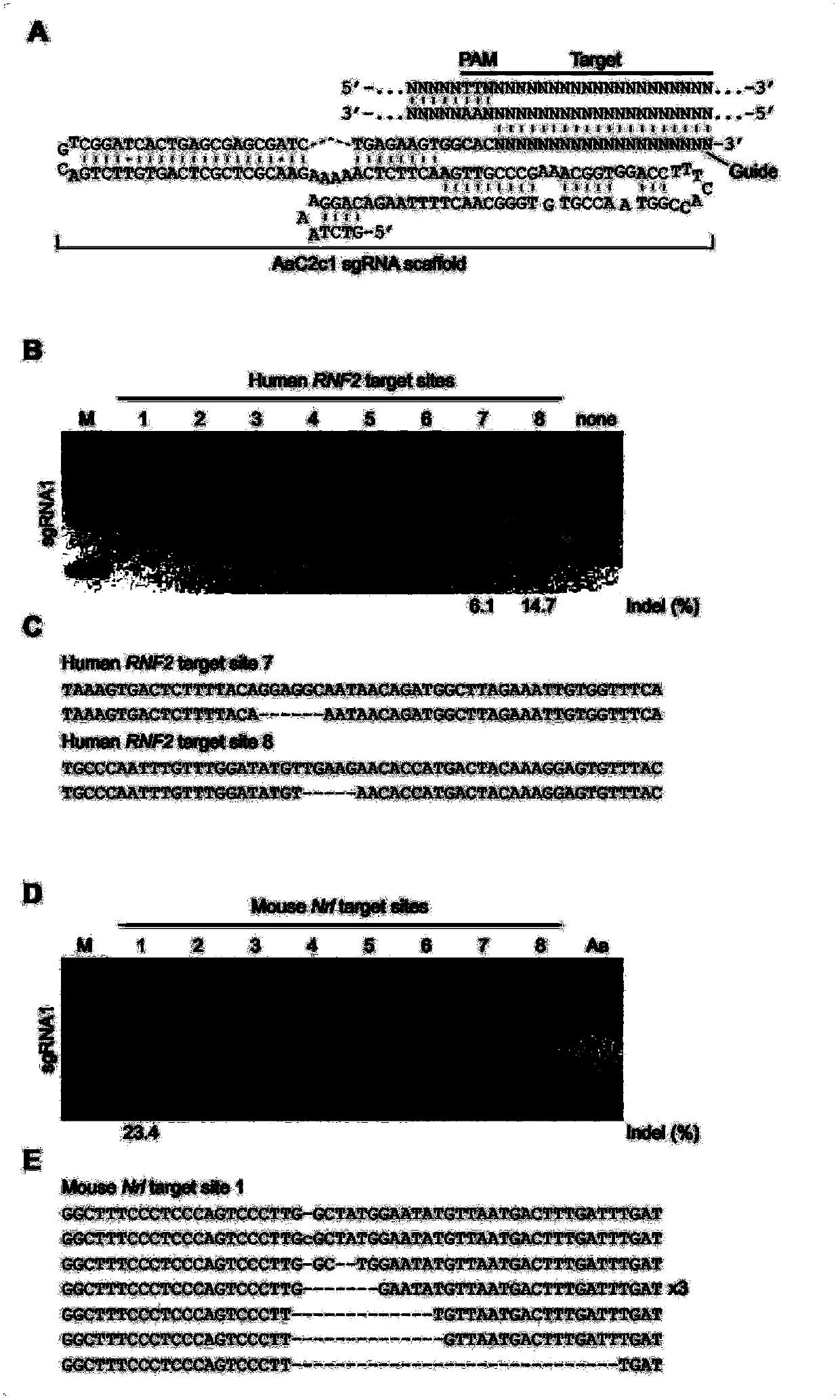
[0160] image 3 A is a schematic diagram of the AaC2c1 sgRNA-DNA-targeting complex.
[0161] image 3 B shows T7EI analysis of indels generated at human RNF2 target sites. Numbers under lanes with mutations show Indel ratios. Triangles indicate cut fragments.
[0162] image 3 C shows from image 3 Sanger sequencing results of the cleavage products of B. Red font highlights PAM sequences.
[0163] T7EI experiments showed that AaC2c1 induced an indel at the mouse Nr1 locus ( image 3 D). image 3 E shows from image 3 Allelic sequence resulting from cleavage of target 1 of the Nrl gene of D.
[0164] Thus, AaC2c1 mediates robust genome editing in mammalian cells. Figure 4 The data further confirmed this conclusion. Figure 4 A AaC2c1 induces a...
Embodiment 3
[0170] Example 3, sgRNA optimization
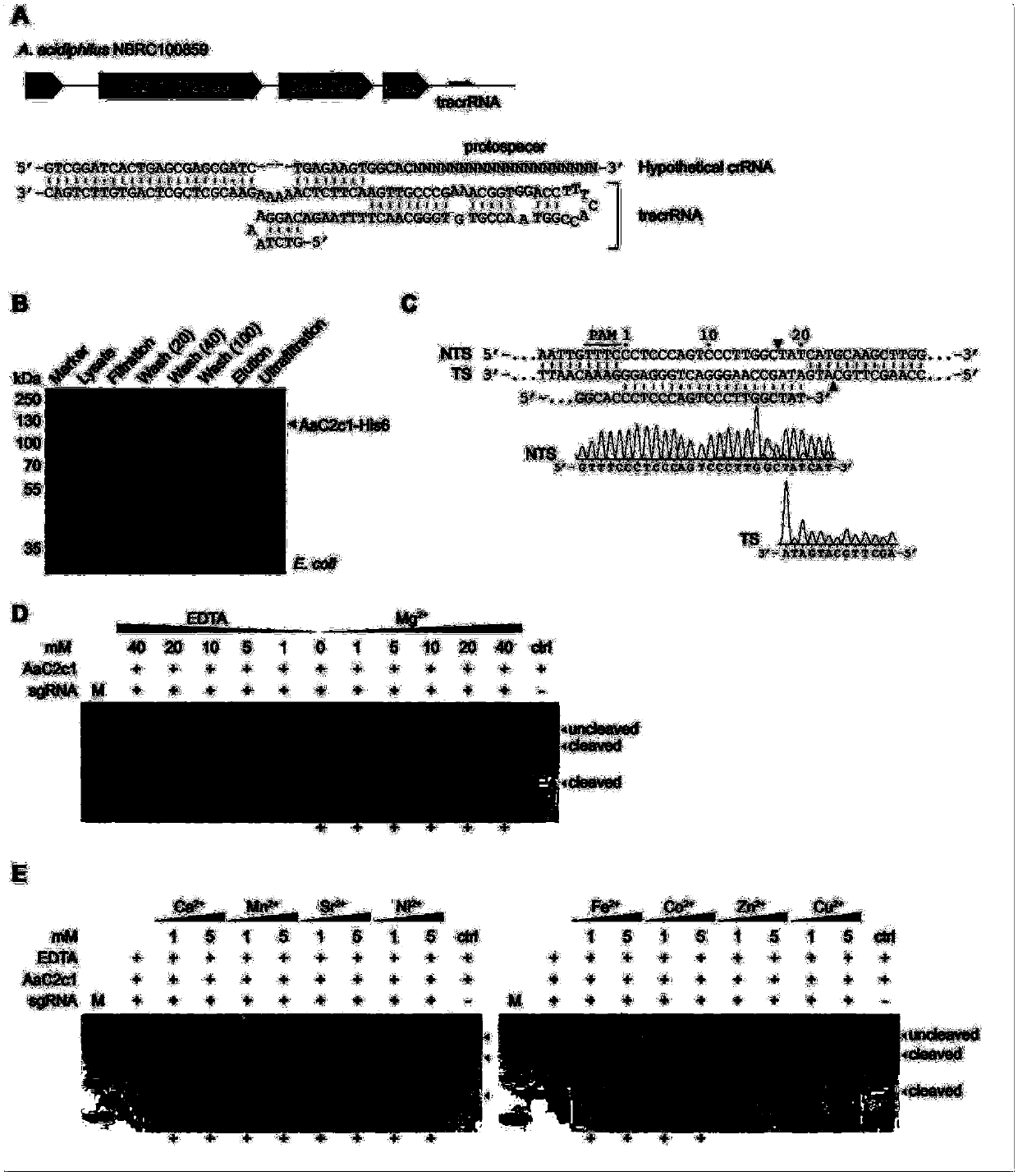
[0171] This example optimizes the single guide RNA (sgRNA) that guides AaC2c1 genome editing. The original sgRNA was sgRNA1 constructed based on the tracrRNA in the AaC2c1 locus and the putative crRNA of A. acidoterrestris.
[0172] Image 6 A shows the structure of different versions of the sgRNA scaffold sequence with 5' truncated at stem-loop 3 of sgRNA1. Figure 5 A shows that truncation and disruption of stem-loop 3 of the sgRNA abolished the targeting activity of AaC2c1 to target site 8 of the human endogenous gene RNF2 in vivo. Image 6 B shows that truncation and disruption of stem-loop 3 of the sgRNA abolished AaC2c1 targeting activity in vitro. Image 6 C shows that truncation and disruption of stem-loop 3 of the sgRNA abrogates the targeting activity of AaC2c1 to target site 1 of the mouse endogenous gene Nrl in vivo.
[0173] Image 6 D shows the structure of different versions of the sgRNA scaffold sequence truncated and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com