CircRNA-disease association predicating method based on network model
A prediction method and network model technology, applied in the interdisciplinary fields of bioinformatics and artificial intelligence, can solve problems such as time-consuming, waste of manpower and financial resources, and achieve the effects of short time-consuming, high prediction accuracy and cost reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
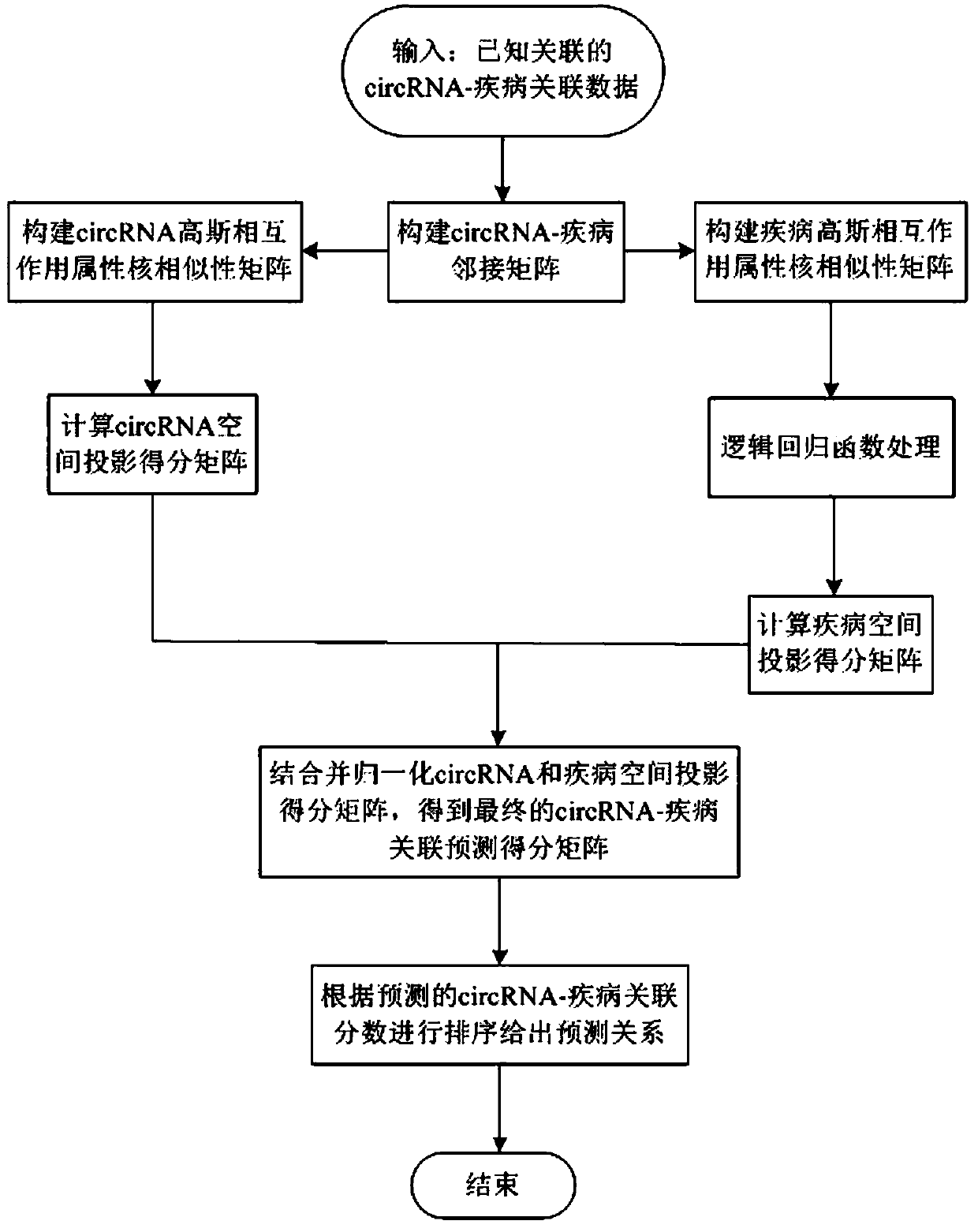
[0036] refer to figure 1 , a network model-based circRNA-disease association prediction method, including the following steps:
[0037] 1) Obtain the circRNA-disease association data set and construct the adjacency matrix A about the circRNA-disease association: Obtain the circRNA-disease association data confirmed by biological experiments from the circRNADisease database. Due to the small amount of chicken and mouse species association data, it is not representative , we deleted the circRNA-disease association data of chicken and mouse species, and only kept the circRNA-disease association data of human species, and finally obtained 239 pairs of different circRNA and disease association data, involving 34 types of diseases and 223 types of circRNAs. Define D={d(1), d(2), d(3),...,d(nd)} to record the set of nd diseases, C={c(1), c(2), c (3),..., c(nc)} to record the collection of nc circRNAs, and build an adjacency matrix A nd×nc Indicates the relationship between circRNA ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com