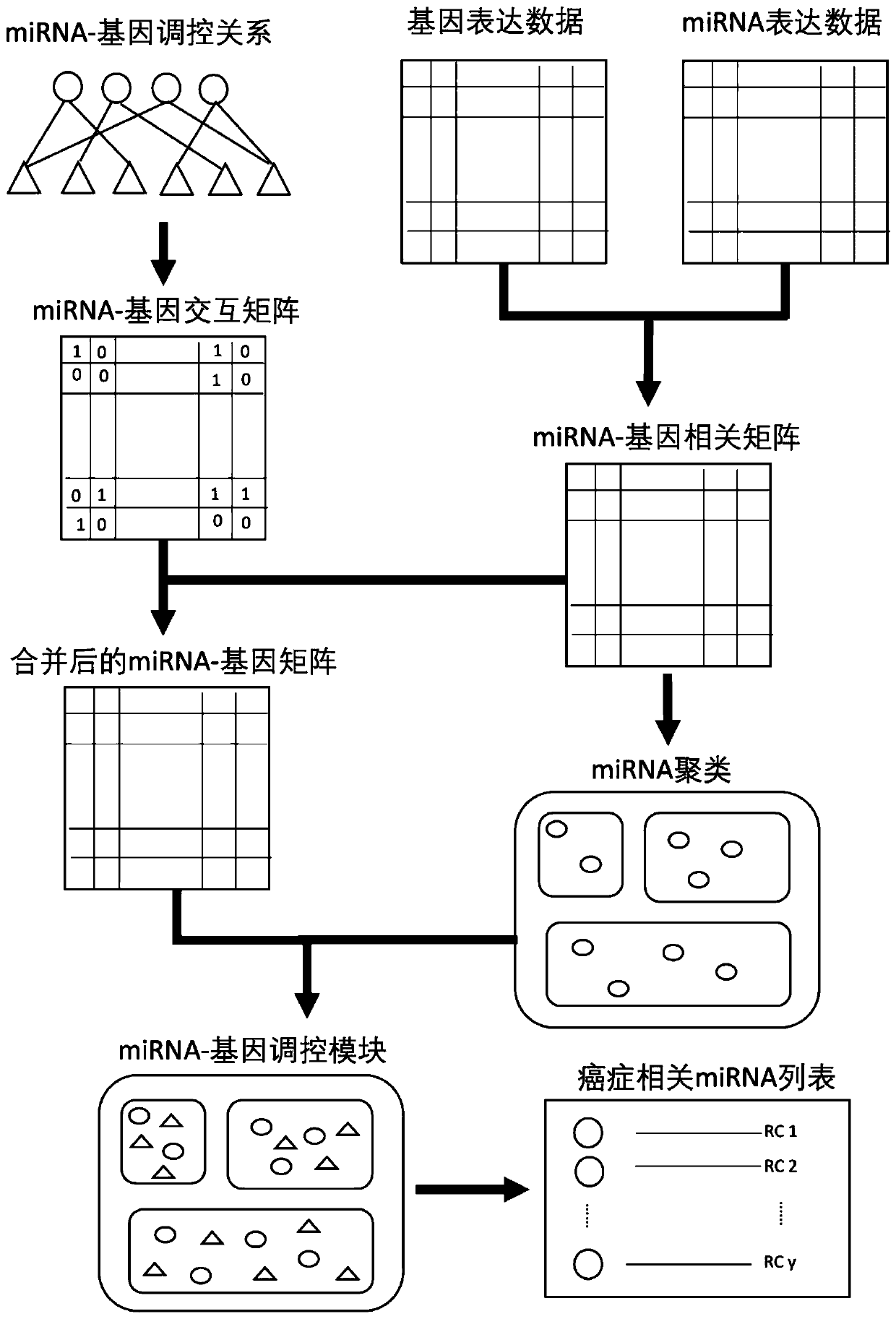
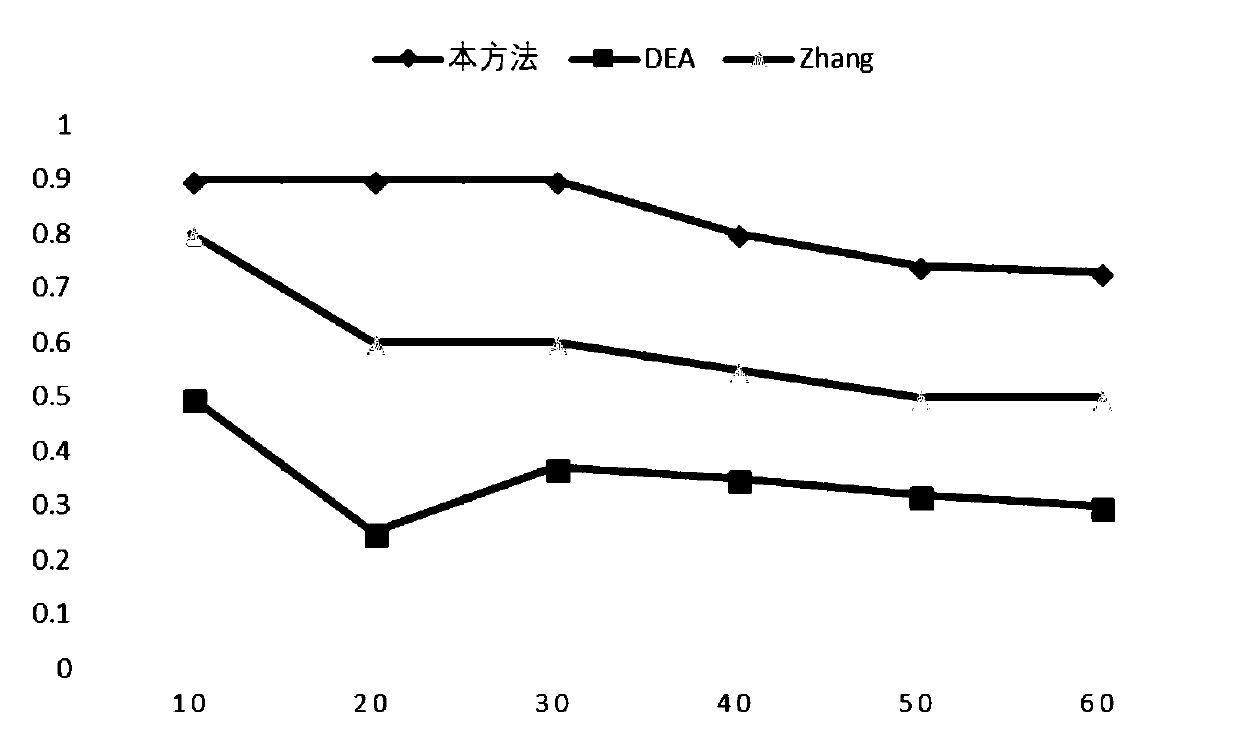
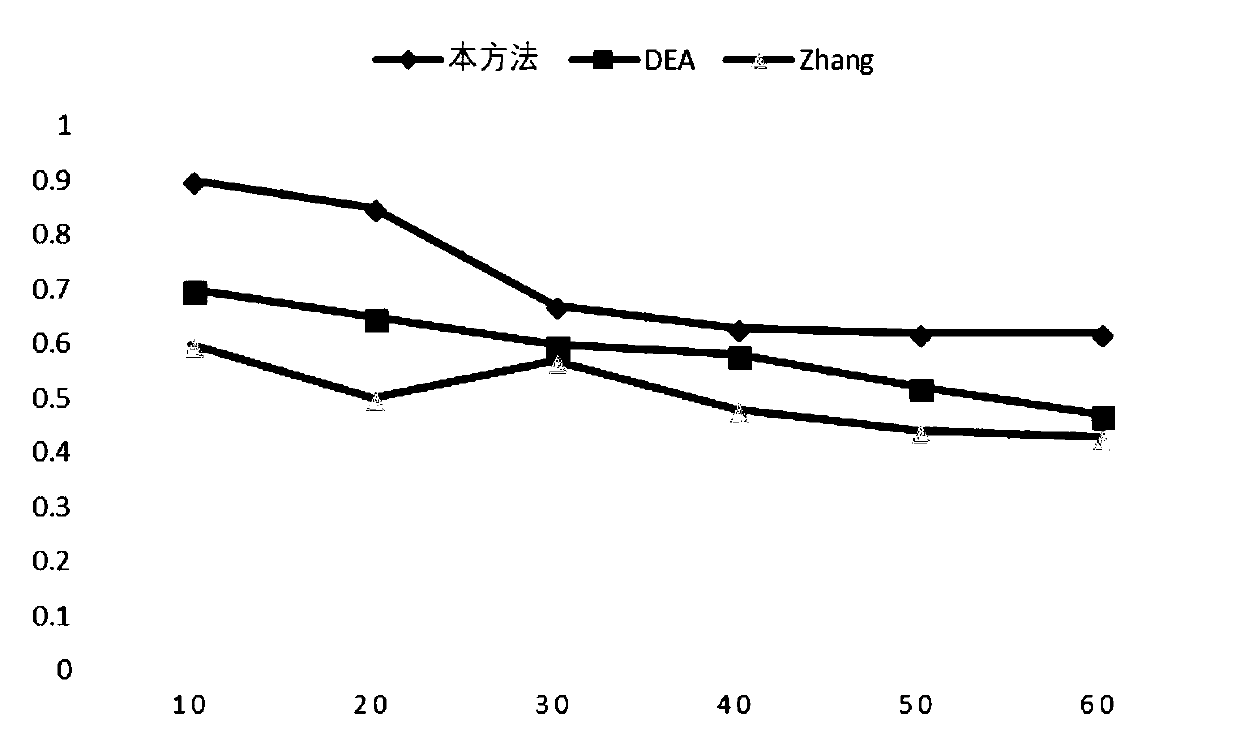
Cancer-related MicroRNA identification method based on miRNA-gene regulation module
A cancer-related and identification method technology, applied in the field of data mining, can solve the problem of not fully considering the miRNA regulatory relationship, and achieve the effect of improving efficiency and accuracy, and reducing the scope of calculation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0032] In order to make the object, technical solution and advantages of the present invention clearer, the present invention will be further described in detail in combination with experiments below. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
[0033] The hardware environment is mainly a PC host. Among them, the CPU of the PC host is Intel(R) Core(TM) i7-6700, 3.40GHz, the memory is 32GB RAM, and the 64-bit operating system. The software uses Windows 7 as the platform and is implemented in R language in the RStudio environment. The RStudio version is 1.1.142 and the R language version is 3.5.0.
[0034] The data used are three relatively complete cancer data sets downloaded from TCGA, breast cancer (Breast Invasive Carcinoma, BRCA), prostate cancer (Prostate Adenocarcinoma, PRAD), lung adenocarcinoma (LungAdenocarcinoma, LUAD), these three cancer-related There are r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com