Model for predicting prognosis of lung squamous cell carcinoma with seven genes as biomarkers, and establishing method thereof
A lung squamous cell carcinoma and prognosis technology, applied in the fields of genetic technology and biomedicine, can solve the problems of insensitive and accurate prognosis of lung squamous cell carcinoma, low overall survival rate, and no reference standard for prognosis judgment.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 contains seven genes as biomarkers to predict the prognosis of lung squamous cell carcinoma model building method.
[0032] 1. Data collection.
[0033] A total of 551 samples were obtained from The Cancer Genome Atlas (TCGA) database (http: / / cancergenome.nih.gov / ), including 49 normal samples and 502 lung squamous carcinoma samples. Meanwhile, all samples contained corresponding clinical data on age, sex, race, smoking status, cancer stage, survival time and RNA expression profile from the database. Differentially expressed genes were screened by R (http: / / www.r-project.org) meeting the selection criteria as follows: 1) p 1.
[0034] 2. Construction of Cox regression model.
[0035] By collecting differently expressed genes, 363 samples were randomly selected as the training set, and 188 samples were used as the R-based test set; Cox univariate analysis was used to obtain prognosis-related genes. Cox multivariate analysis was performed by stepwise regressi...
Embodiment 2
[0044] Example 2 Application of a model containing seven genes as biomarkers to predict the prognosis of lung squamous cell carcinoma.
[0045] 1. Cox regression model predicts lung squamous cell carcinoma.
[0046] The 7 genes collected from the Cox multivariate regression analysis and the risk score formula can be calculated in each sample list as follows:
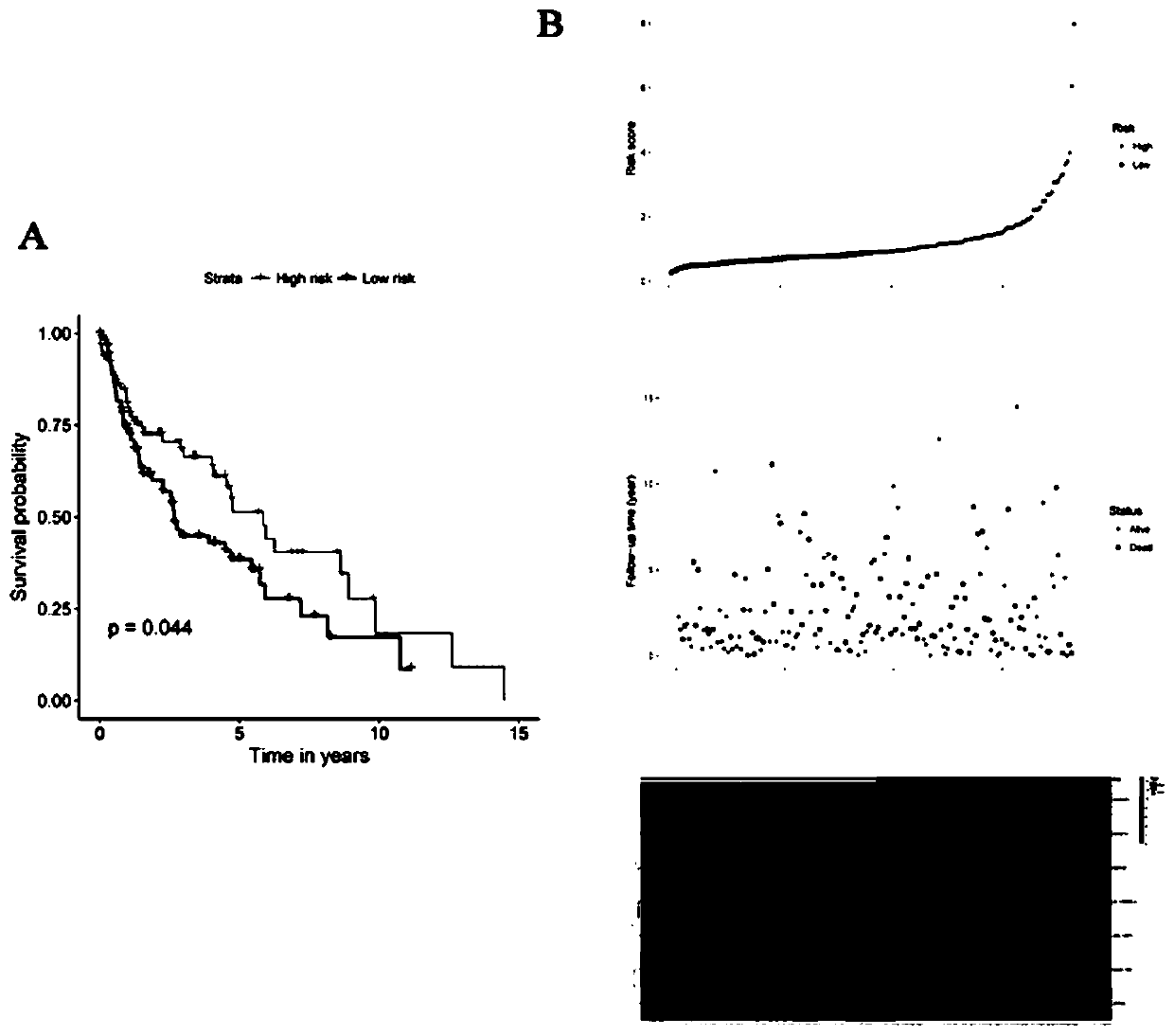
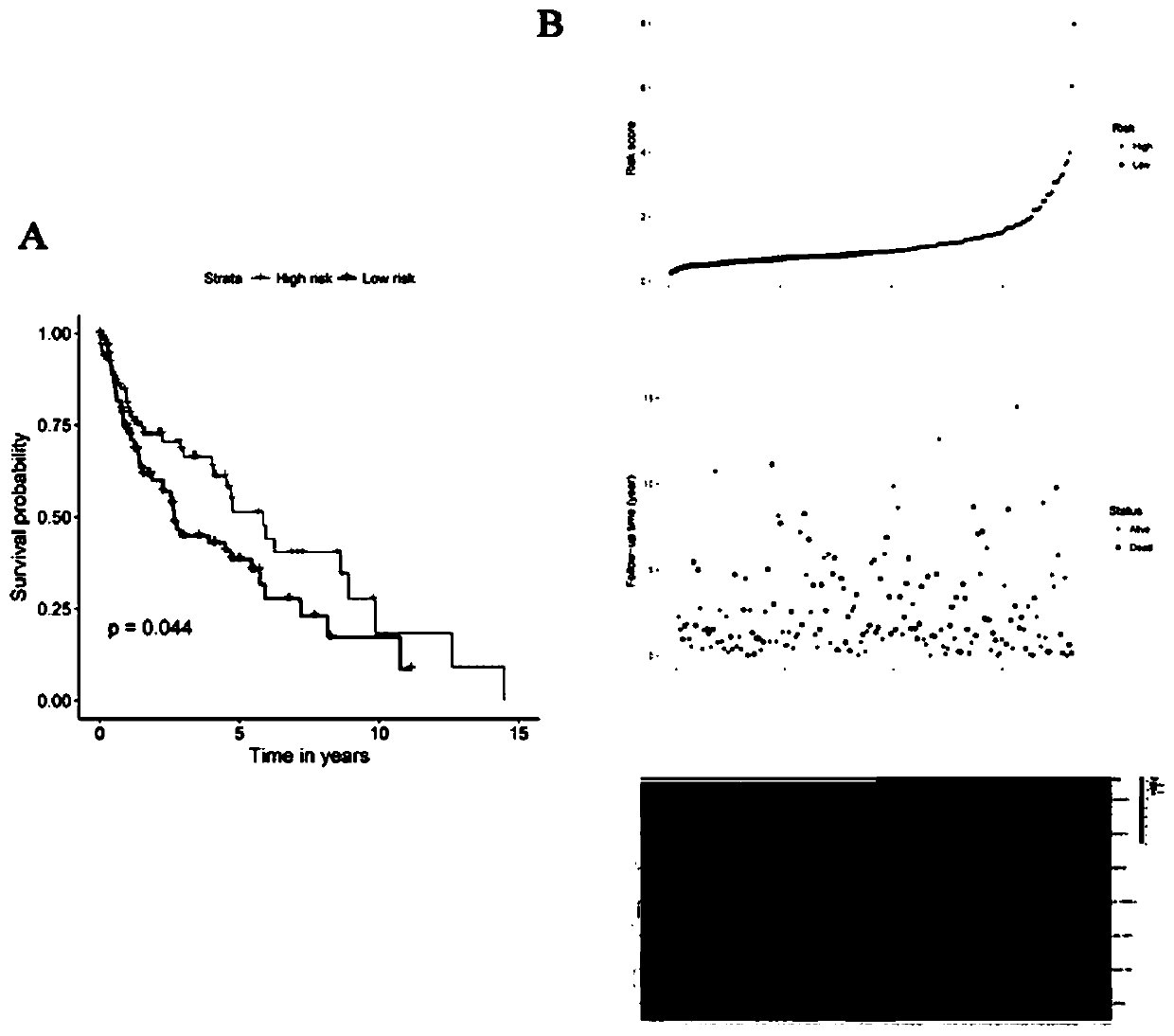
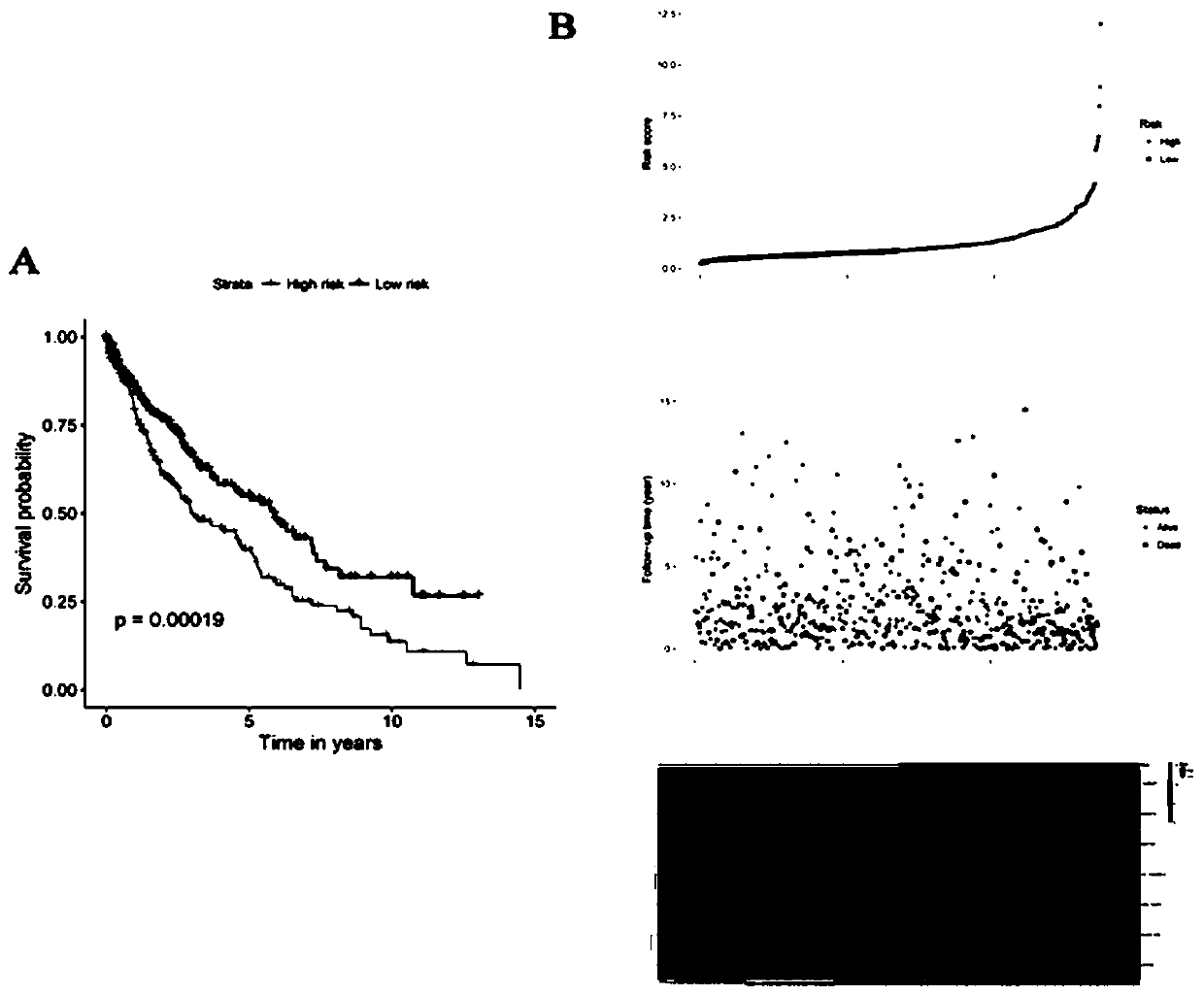
[0047] Risk score = -0.1311*CSRNP1+0.1390*MIR27A-0.1951*CLEC18B+0.1708*AC130456.4+0.1702*DEFA6+0.1821*ARL14EPL+0.1135*ZFP42. Only CSRNP1 and CLEC18B were positively associated with survival and acted as protective factors. MIR27A, AC130456.4, DEFA6, ARL14EPL and ZFP42 were inversely associated with survival and played a role in increased risk (Table 1).
[0048]
[0049] 2. Performance of the training set risk score.
[0050] According to the median risk score as the threshold, lung squamous cell carcinoma samples were divided into high-risk group and low-risk group. Kaplan-Meier curves were used to indicate diffe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com