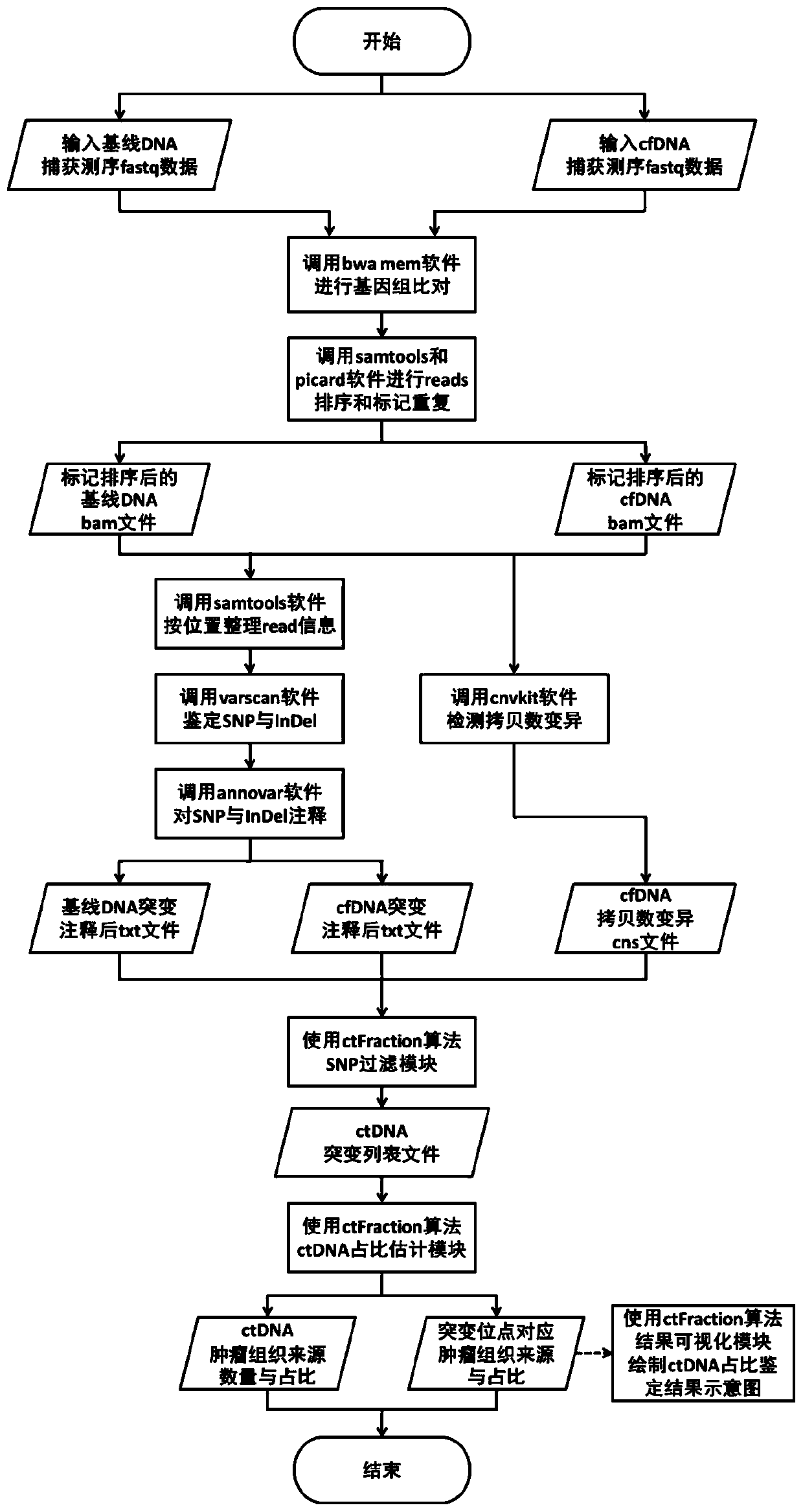
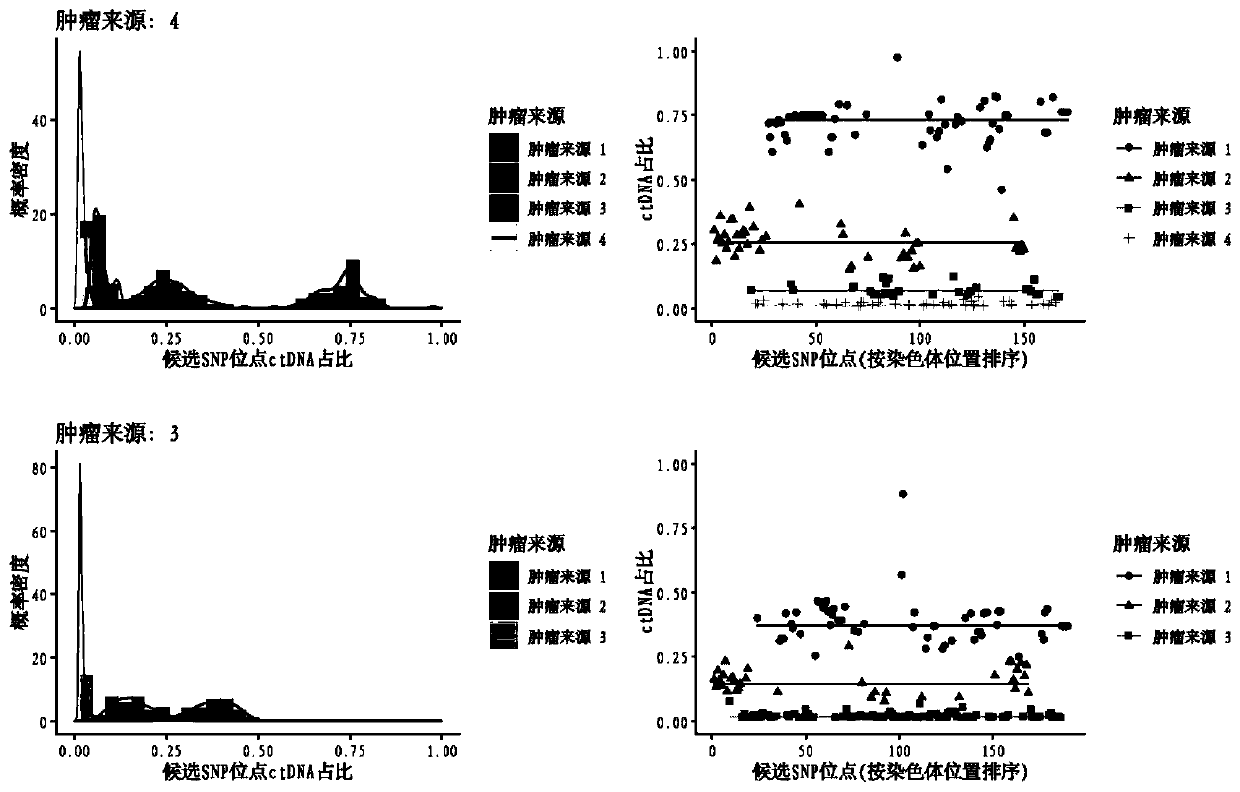
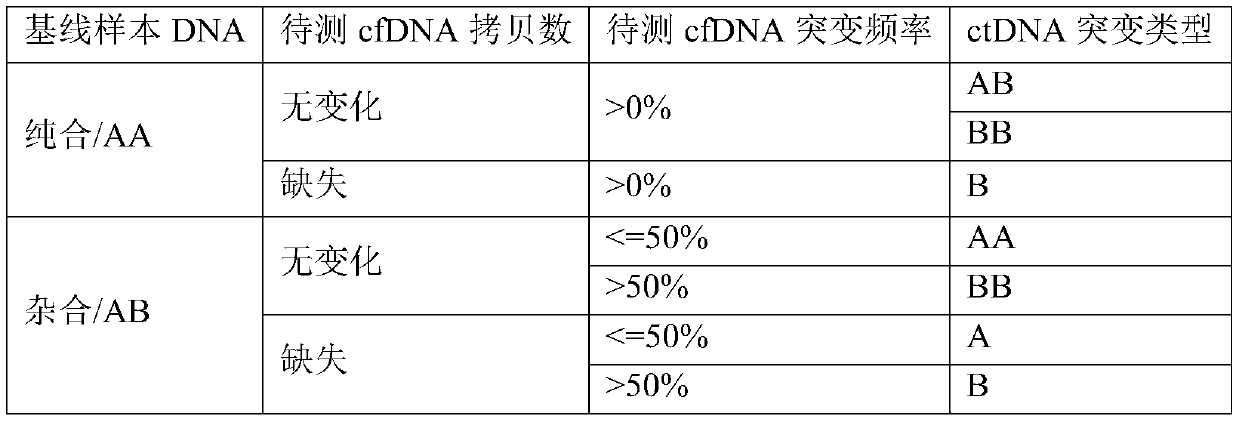
Detection method and detection device for ctDNA proportion based on capture sequencing
A detection method and sequencing technology, applied in the field of genetic engineering, can solve the problem of low accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0178] 1. Sample Preparation
[0179] Select blood cell DNA and cfDNA samples from 6 tumor patients (respectively denoted as sample A, sample B, sample C, sample D, sample E, and sample F), and perform the following operations respectively:
[0180] 1) Purification and quantification of cfDNA
[0181] The cfDNA in the sample to be tested will be subjected to 2100 quality inspection to see if there are large fragments. For the purification of cfDNA samples containing large fragments, since the magnetic beads have the characteristic of first adsorbing large fragments, first use 0.5 times magnetic beads to purify, at this time, large fragments are adsorbed on the magnetic beads, and the supernatant is sucked into 2.5 times magnetic beads, All products except large fragments were recovered. Qubit quantification was performed on all samples.
[0182] 2) Purification and quantification of blood cell DNA
[0183] Take a sufficient amount of 6 blood cell DNA in the sample to be te...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com