Molecular classification and application of glioma based on cdc20 gene co-expression network
A CDC20-M, gene expression technology, applied in the biological field, can solve the problems of chromosomal instability, genome instability, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
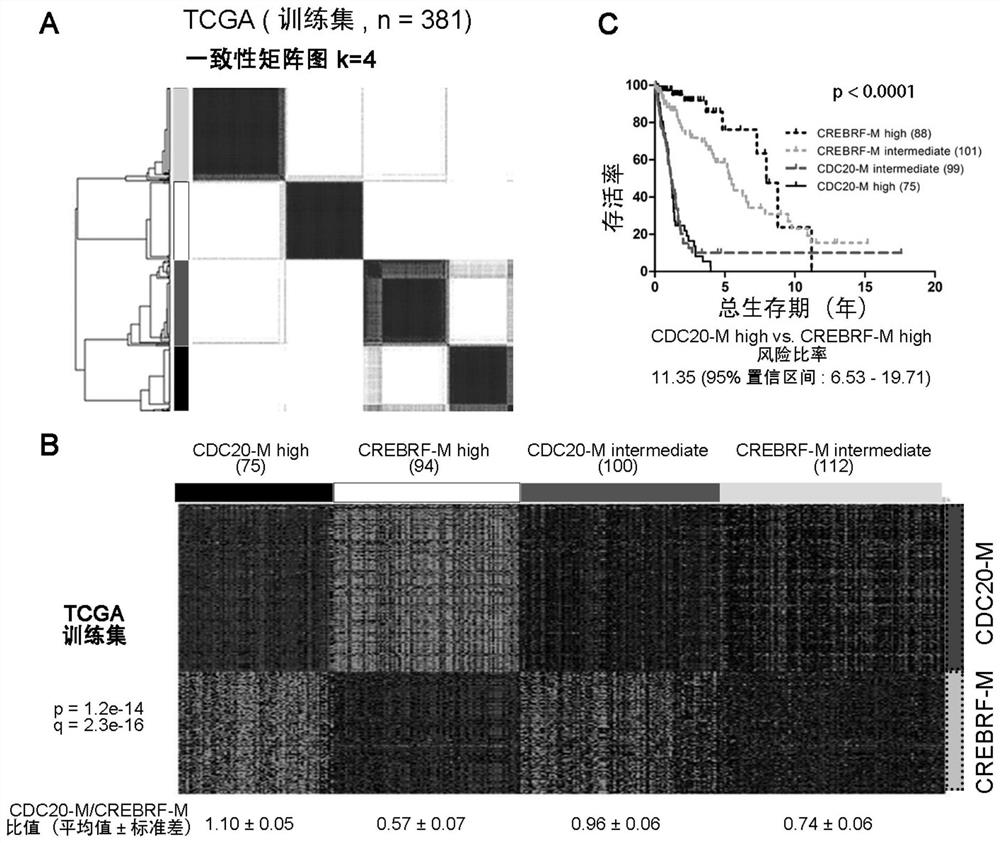
[0053] Example 1, the establishment of CDC20-M and CREBRF-M gene co-expression modules
[0054] The genomic instability of tumors is manifested in the fact that tumor samples carry a large number of dynamically changing chromosomal abnormalities and gene mutations. Different tumor samples may carry different chromosomal abnormalities and gene mutations, and tumors may accumulate important genomic abnormalities at a certain developmental point, and then their genomes remain relatively stable. Therefore, how to identify tumor samples with unstable genomes has become a key issue in this field. One possible strategy is to screen gene expression profiles that correlate with the number of chromosomal variants in representative samples.
[0055]In order to screen specific gene groups that are closely correlated with genome instability in glioma, the present invention uses Pearson correlation analysis, and takes Pearson correlation coefficient (Pearson correlation coefficient, PCC)>7...
Embodiment 2
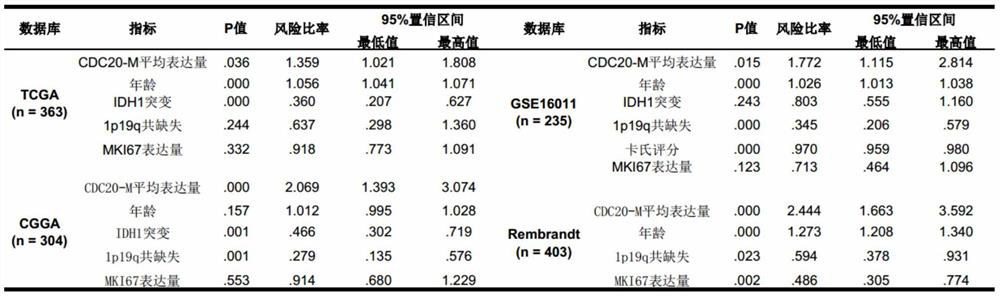
[0058] Example 2, Comparison of CDC20-M and existing glioma prognostic markers
[0059] In order to verify the role of CDC20-M in the occurrence and development of glioma, the present invention first analyzed the ability of CDC20-M in the prognosis diagnosis and prediction of glioma patients, and explored whether CDC20-M has all types and grades of glioma The ability to distinguish tumors according to their prognostic differences. For the prognosis diagnosis of glioma, there are currently some commonly used markers in clinic, such as age, IDH1 mutation, co-deletion of chromosome arm 1p and 19q, and protein expression level of Ki-67, so the present invention combines CDC20-M with this Several mature prognostic markers were compared by multivariate cox regression analysis. The database used covered a total of 1305 patient samples from Europe, the United States and China (Rembrandt, GSE16011, TCGA and CGGA databases). The results are shown in Table 1 shown. It can be found that...
Embodiment 3
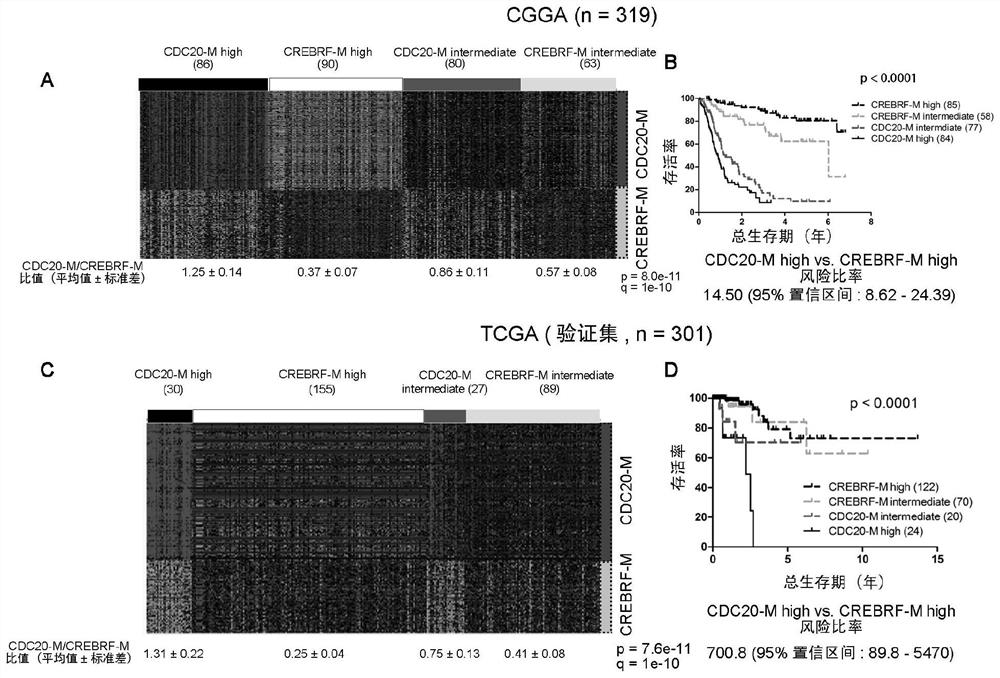
[0073] Example 3, independent sample prediction based on CDC20-M and CREBRF-M
[0074] After the stable CDC20-M grouping is obtained, the present invention uses the independent sample predictive analysis method to group glioma patient samples from other sources based on the TCGA training set. In the TCGA training set, for each group classified by CDC20-M, the average expression of each member in CDC20-M and CREBRF-M is calculated separately, so there is a set of 139+120 members for each group For the average expression level, four groups have four sets of average expression levels, and each set of average expression levels is called a group-specific expression pattern (Centroid). There are four Centroids in four groups. The present invention subsequently obtained 319 cases of glioma samples from CGGA (140 cases of glioblastoma, 67 cases of astrocytoma, 40 cases of oligodendroglioma, 72 cases of oligoastrocytoma) and New TCGA sample of 301 low-grade gliomas (100 astrocytomas,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com