Screening method of molecular reverse probes for capturing specific region of enriched genome
A molecular reverse probe, a specific region technology, applied in the field of genetic engineering probes, which can solve the problems of the optional range of probes and the less consideration of experimental characteristics.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] Embodiment 1: detect the MIP probe design of human gene BRCA1 / 2 exon region
[0076] Using the method of the present invention, the molecular reverse probe design of the single-molecule tag marking is carried out on the coding region and the cut region of the BRCA1 and BRCA2 genes in the human genome. The coverage of this probe design for targeted capture was counted, and some of the probes were experimentally verified, and the uniformity and specificity of the sequencing data were systematically evaluated.
[0077] The specific design process is as follows:
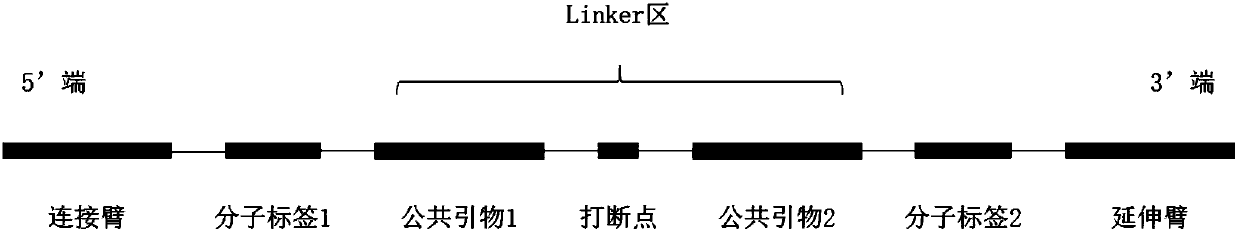
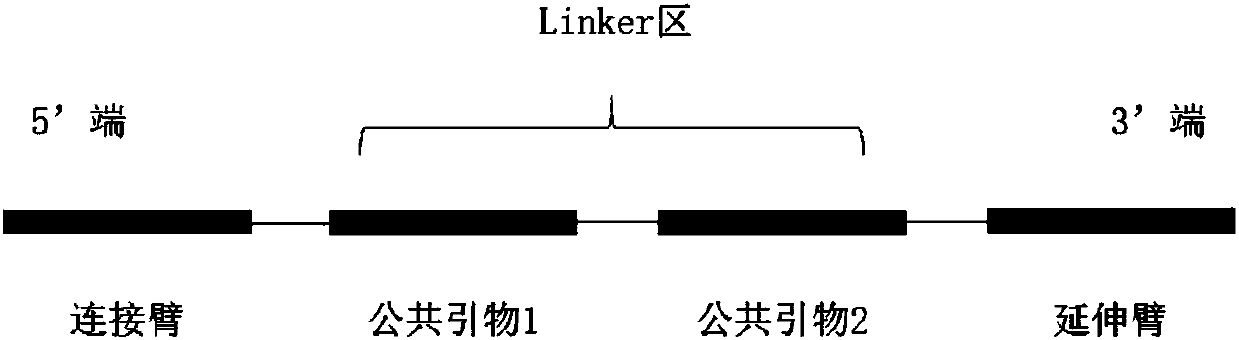
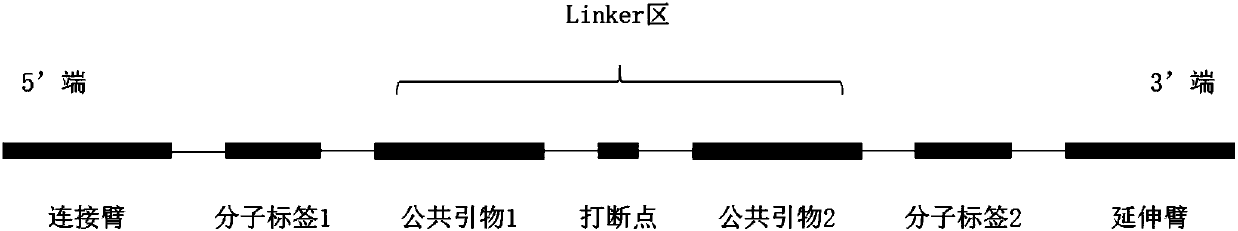
[0078] 1. Selection of probe structure:
[0079] In this experimental design, the expected detection target is 1% of Somatic mutations, so UIDs are introduced into the probe structure, and the UIDs are distributed on both sides of the linker region. 6bp random bases were used each time, and the 2bp near the tether or extension arm were designed as random bases mutually exclusive with the reference genotype.
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com