Method and device for predicting ribonucleic acid folding structure based on extended structure with pseudoknots
A technology of extended structure and ribonucleic acid, which is used in the analysis of two-dimensional or three-dimensional molecular structures, bioinformatics, instruments, etc., and can solve problems such as low accuracy of model parameters, low parameter accuracy, and large errors in calculation methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
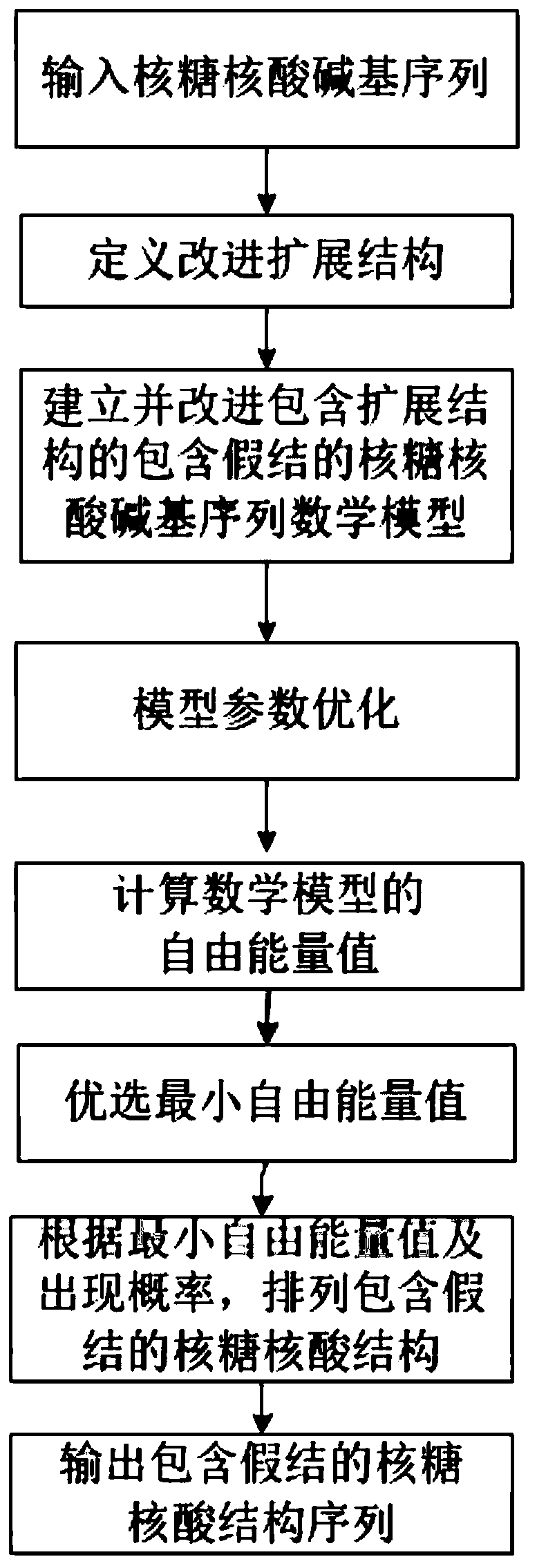
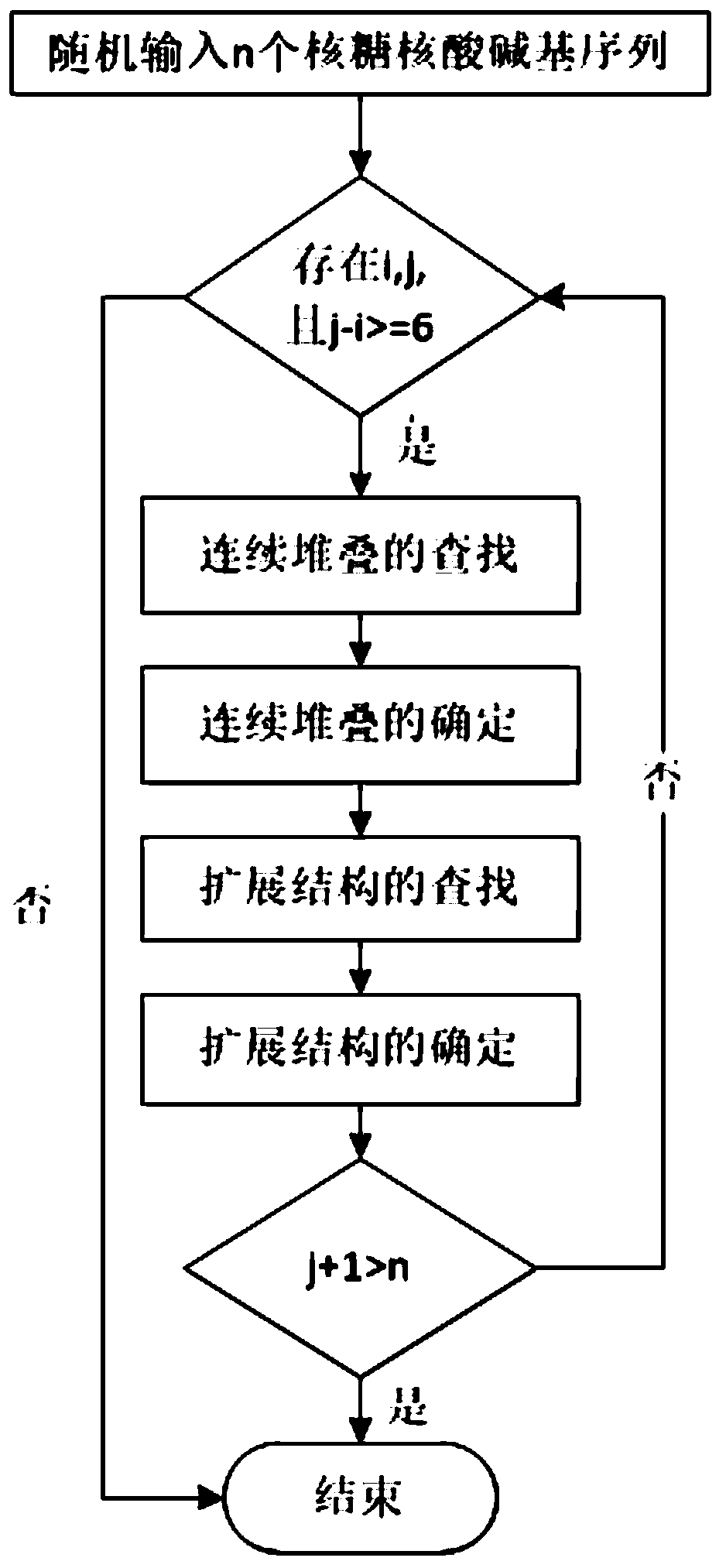
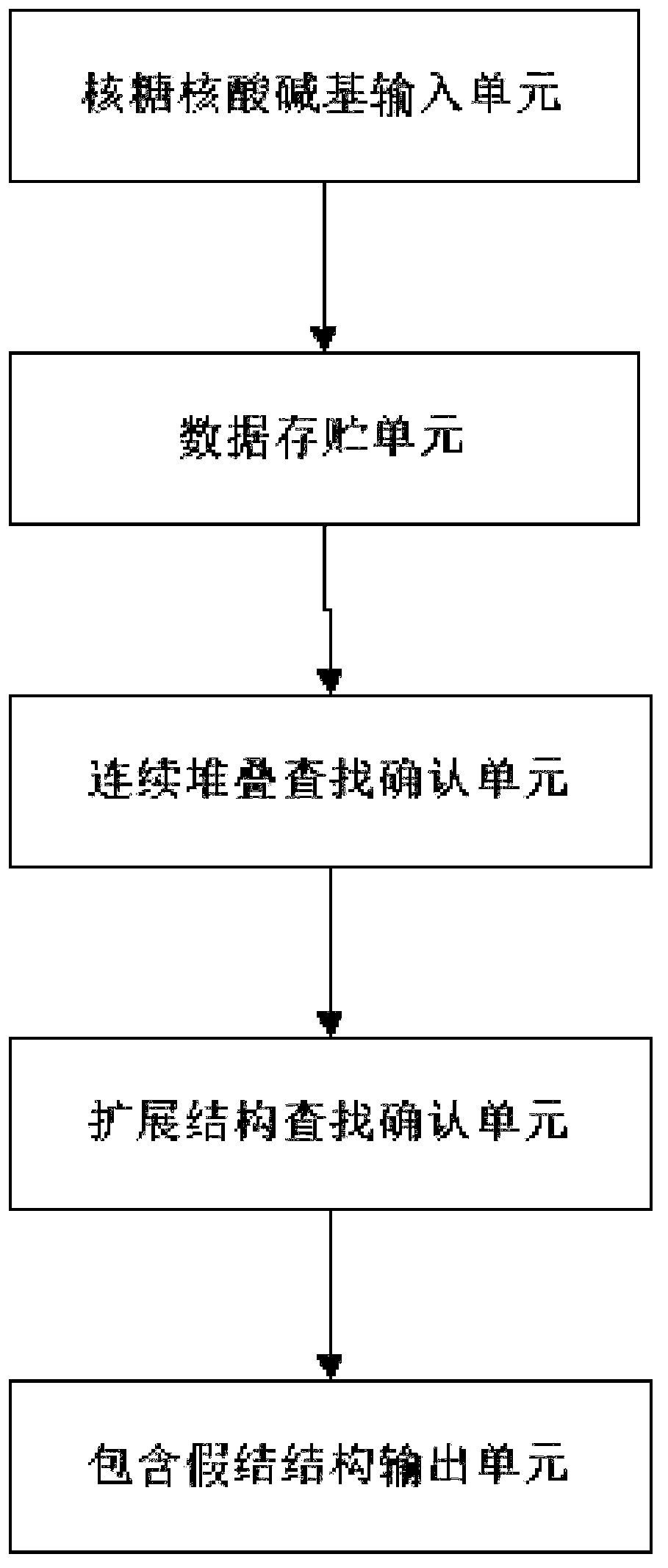
[0039] First, the concepts of RNA sequences, base pairs, pseudoknots, etc. are explained.
[0040] RNA sequence: the arrangement order of the four bases on the side chain of the RNA molecule, generally using A, U, G, C to represent the base pair: if s i ·s j ∈{AU,CG,GU}, then s i ·s j form base pairs. The energy of base pair stacking is negative. false knot: if s i ·s j ∈{AU,CG,GU},s k ·s l ∈{AU,CG,GU}, ii ·s j with s k ·s l form a false knot.
[0041] RNA primary structure: the arrangement order of the four bases on the side chain of the RNA sequence. In general RNA sequences end from 5' to 3', so the whole sequence s is denoted as s=s 1 the s 2 …s n ,s i is the i-th base of the RNA sequence, s i ∈{A,U,G,C}, RNA base subsequence s i,j is a sequence fragment of s, denoted as: s i,j =s i …s j .
[0042] RNA secondary structure: a set of base pairs in the RNA sequence constitutes the RNA folding structure, denoted by S. For any base pair, if s i ·s j ∈ S...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com