LAMP primer combination for detecting 4 gram-negative bacteria in intraocular fluid and application thereof
A primer combination and primer set technology, applied in the biological field, can solve the problems of long PCR detection time, easy contamination, and application limitations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0133] Embodiment 1, the preparation of kit
[0134] The kit consists of four LAMP primer sets, each for the detection of 1 Gram-negative bacterium in intraocular fluid.
[0135] The primer set used to detect E. coli is as follows (5'→3'):
[0136] Outer primer F3 (SEQ ID NO: 1): GGCATCGTGGTGATTGATGA;
[0137] Outer primer B3 (SEQ ID NO: 2): GGTTCGTTGGCAATACTCCA;
[0138] Internal primer FIP (SEQ ID NO: 3): TCTTTCGGCTTGTTGCCCGCCTGCTGTCGGCTTTAACCTC;
[0139] Internal primer BIP (SEQ ID NO: 4): TACAGCGAAGAGGCAGTCAACGTTTTGGTTTTTGTCACGCGCTATC;
[0140] Loop primer LF (SEQ ID NO: 5): TTCGAAACCAATGCCTAAAGA;
[0141] Loop primer LB (SEQ ID NO: 6): GCGCACTTACAGGCGATT.
[0142] The primer set used to detect Pseudomonas aeruginosa is as follows (5'→3'):
[0143] Outer primer F3 (SEQ ID NO: 7): CAAggTgTTCATCCACgA;
[0144] Outer primer B3 (SEQ ID NO: 8): CgCTCCAgCgCTTTTCC;
[0145] Internal primer FIP (SEQ ID NO: 9): ACTCgTCgCCCATCTCgATggACTgAACgCCggTAACCA;
[0146] Internal pri...
Embodiment 2
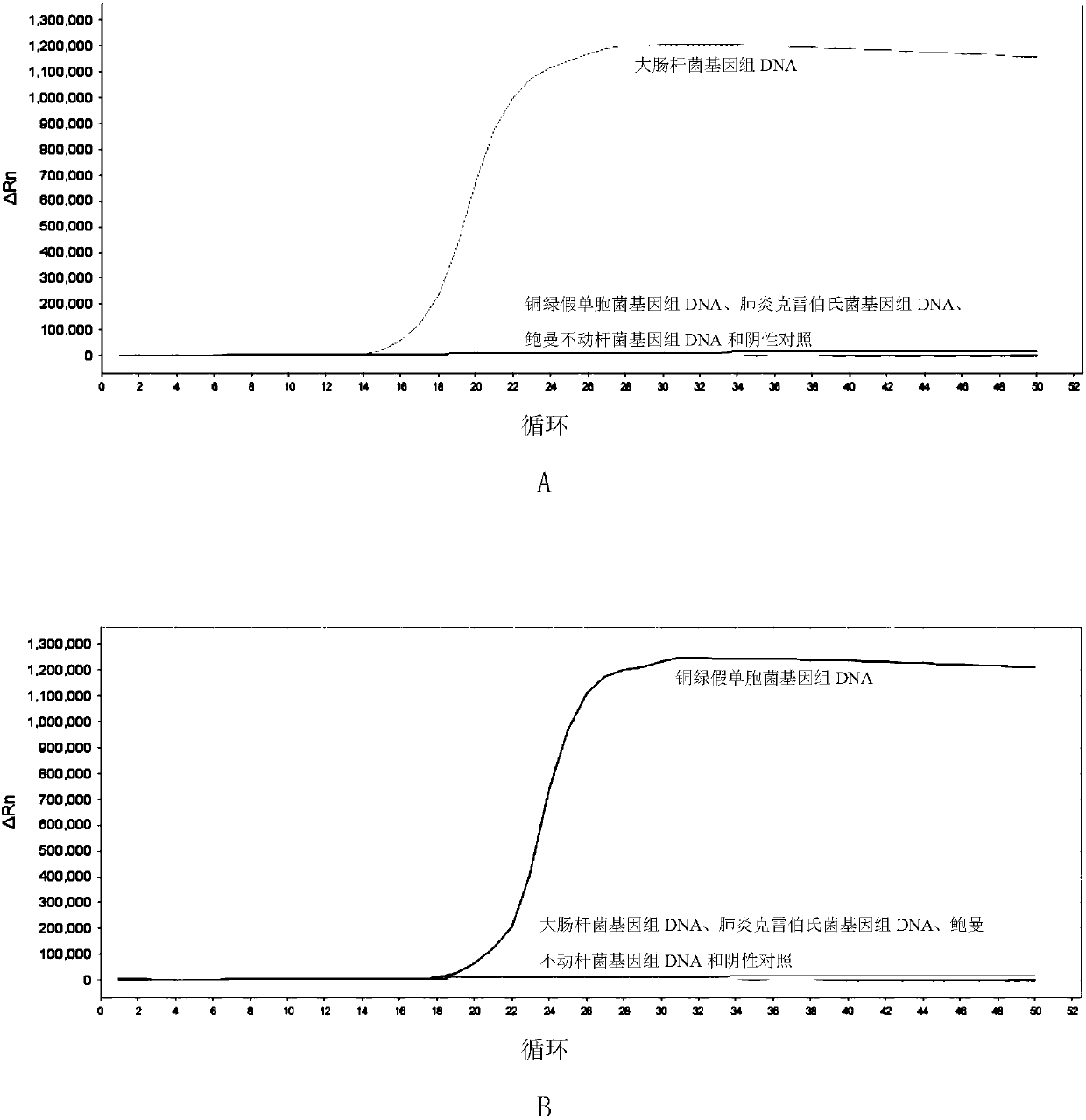
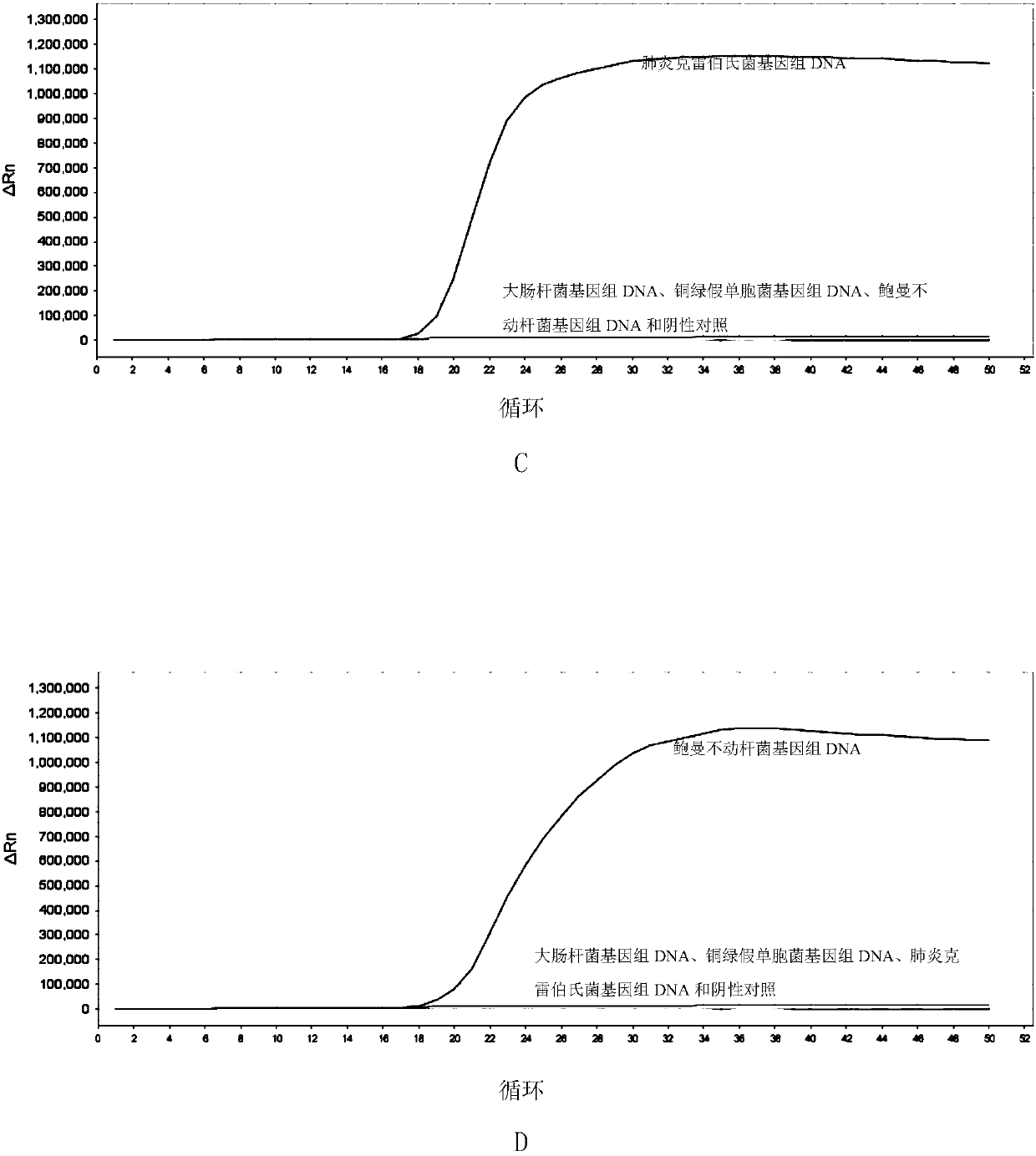
[0164] Embodiment 2, specificity
[0165] Test sample 1: Escherichia coli.
[0166] Test sample 2: Pseudomonas aeruginosa.
[0167] Test sample 3: Klebsiella pneumoniae.
[0168] Test sample 4: Acinetobacter baumannii.
[0169] Each sample to be tested carries out the following steps respectively:
[0170] 1. Extract the genomic DNA of the sample to be tested.
[0171] 2. Using the genomic DNA extracted in step 1 as a template, each primer set prepared in Example 1 was used to perform loop-mediated isothermal amplification.
[0172] Reaction system (10 μL): 1 μL 10×ThermoPol Buffer, 1.6 μL betaine with a concentration of 5M, 0.1 μL BSA aqueous solution with a concentration of 50 mg / mL, and 0.4 μL MgSO with a concentration of 100 mM 4 Aqueous solution, 0.3 μL 20×EvaGreen, 0.15 μL of 100 mM dNTPs (each), 0.4 μL of 8 U / mL Bst DNA polymerase large fragment, genomic DNA of the sample to be tested (between 50 pg and 50 ng), 1 μL Primer mix, rehydrated to 10 μL. The primer mix...
Embodiment 3
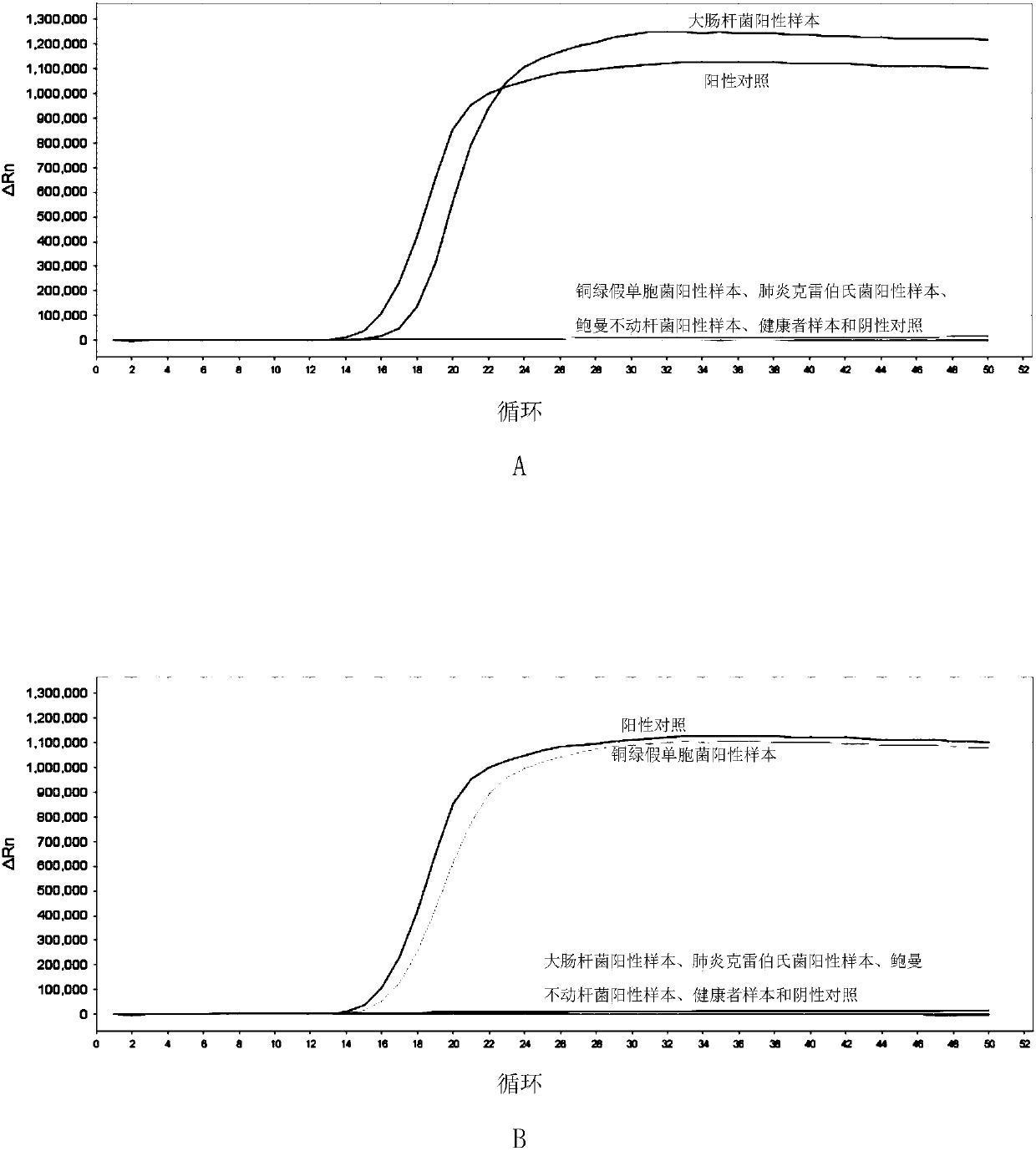
[0181] Embodiment 3, sensitivity
[0182] Test sample 1: Escherichia coli.
[0183] Test sample 2: Pseudomonas aeruginosa.
[0184] Test sample 3: Klebsiella pneumoniae.
[0185] Test sample 4: Acinetobacter baumannii.
[0186] Each sample to be tested carries out the following steps respectively:
[0187] 1. Extract the genomic DNA of the sample to be tested, and perform gradient dilution with sterile water to obtain each dilution.
[0188] 2. Using the dilution obtained in step 1 as a template, the primer sets prepared in Example 1 were used to perform loop-mediated isothermal amplification.
[0189] When the sample to be tested is the sample to be tested 1, the loop-mediated isothermal amplification is performed using primer set I. When the sample to be tested is the sample to be tested 2, loop-mediated isothermal amplification is performed using primer set II. When the sample to be tested is the sample to be tested 3, loop-mediated isothermal amplification is perform...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com