High-sensitive three-dimensional nuclear magnetic resonance spectrum method
A technology of nuclear magnetic resonance spectrum and nuclear magnetic resonance spectrometer, which is applied in the field of highly sensitive three-dimensional nuclear magnetic resonance spectrum, can solve the problems of unrealistic, long time for three-dimensional spectrum sensitivity experiment, and limit the application of three-dimensional nuclear magnetic resonance spectrum, etc., and achieve the effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
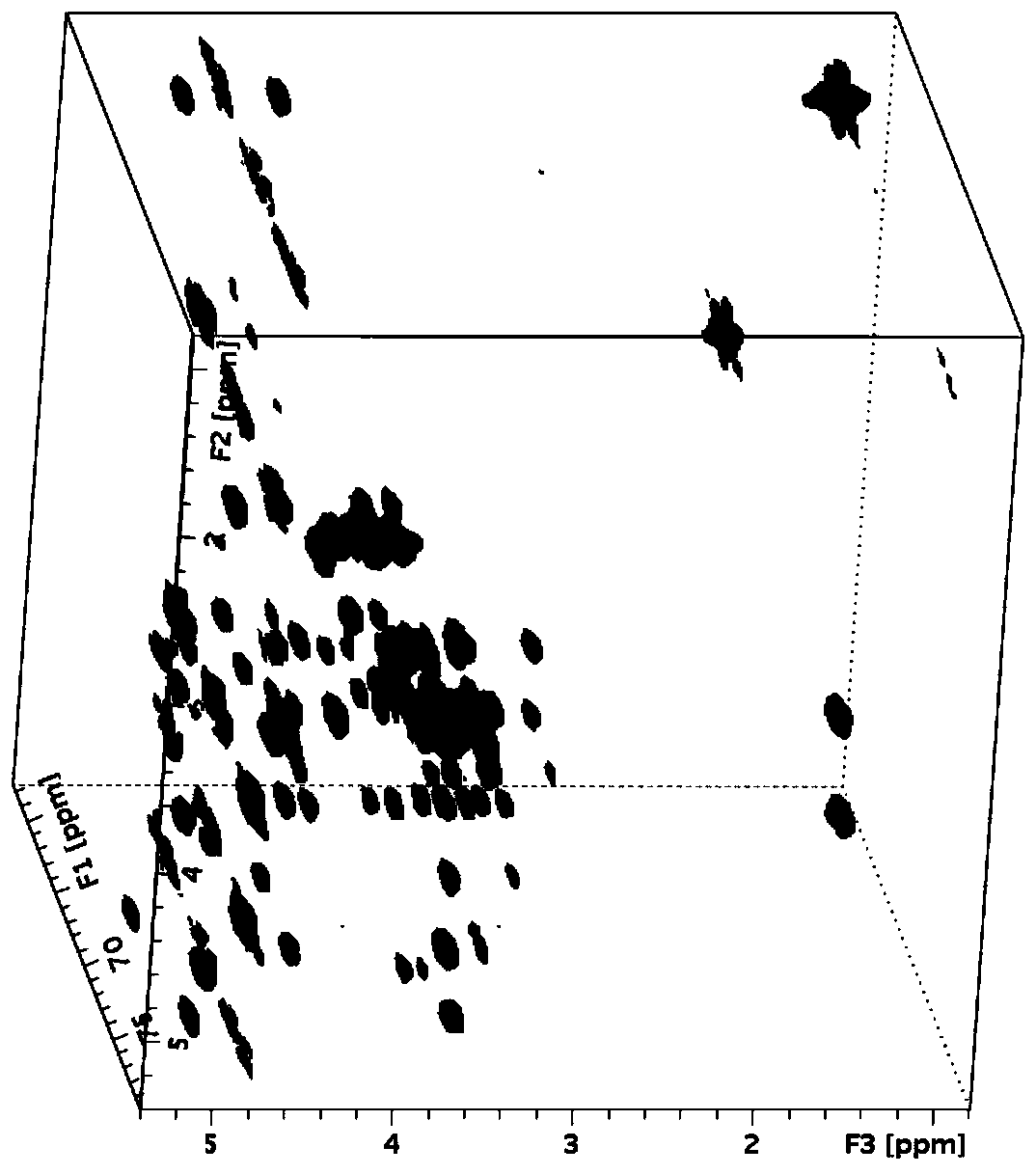
[0032] Example 1: Taking breast milk hexasaccharide as an example to detect the rationality and universality of the method for obtaining a highly sensitive three-dimensional nuclear magnetic resonance spectrum described in the present invention.
[0033] Due to the limitation of resources or extraction and preparation, the sample size of biological oligosaccharides derived from nature will be relatively limited, such as breast milk oligosaccharides. Therefore, in this example, the 2mM heavy lactose hexaose aqueous solution will be used as the sample to detect the three-dimensional HSQC-TOCSY spectrum.
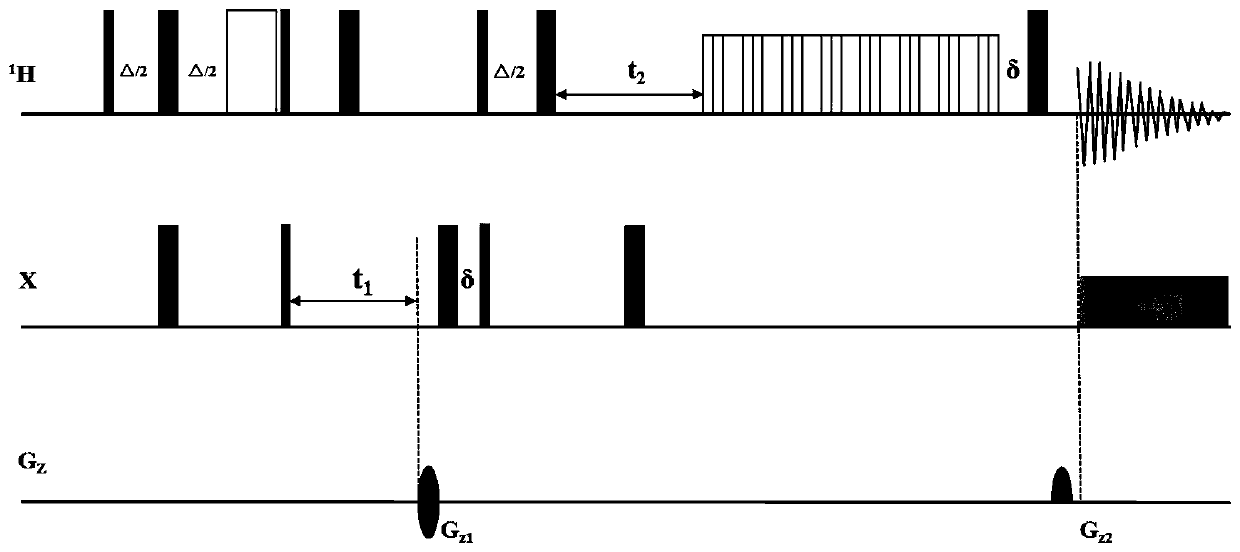
[0034] A method for obtaining high-sensitivity three-dimensional nuclear magnetic resonance spectrum, comprising the following steps:
[0035] 1) Put the sample into a 5mm diameter NMR sample tube, and put the sample tube into a Bruker AVANCE 700MHz NMR spectrometer. The experimental temperature can be selected from a range of 298K, and wait for 10 minutes to allow the temperat...
Embodiment 2
[0041] Example 2: Taking chondroitin sulfate as an example to test the rationality and universality of a method for obtaining a highly sensitive three-dimensional nuclear magnetic resonance spectrum described in the present invention.
[0042] 1) The configuration of the sample to be tested and the optimization of the hydrogen spectrum width are the same as in Example 1.
[0043] 2) The optimization method for the carbon spectrum width is the same as in Example 1, the optimized carbon spectrum width is 30 ppm, and the hydrogen spectrum width is 5.7 ppm.
[0044] 3) Setting the present invention to obtain a highly sensitive three-dimensional HSQC-TOCSY experiment is the same as in Example 1.
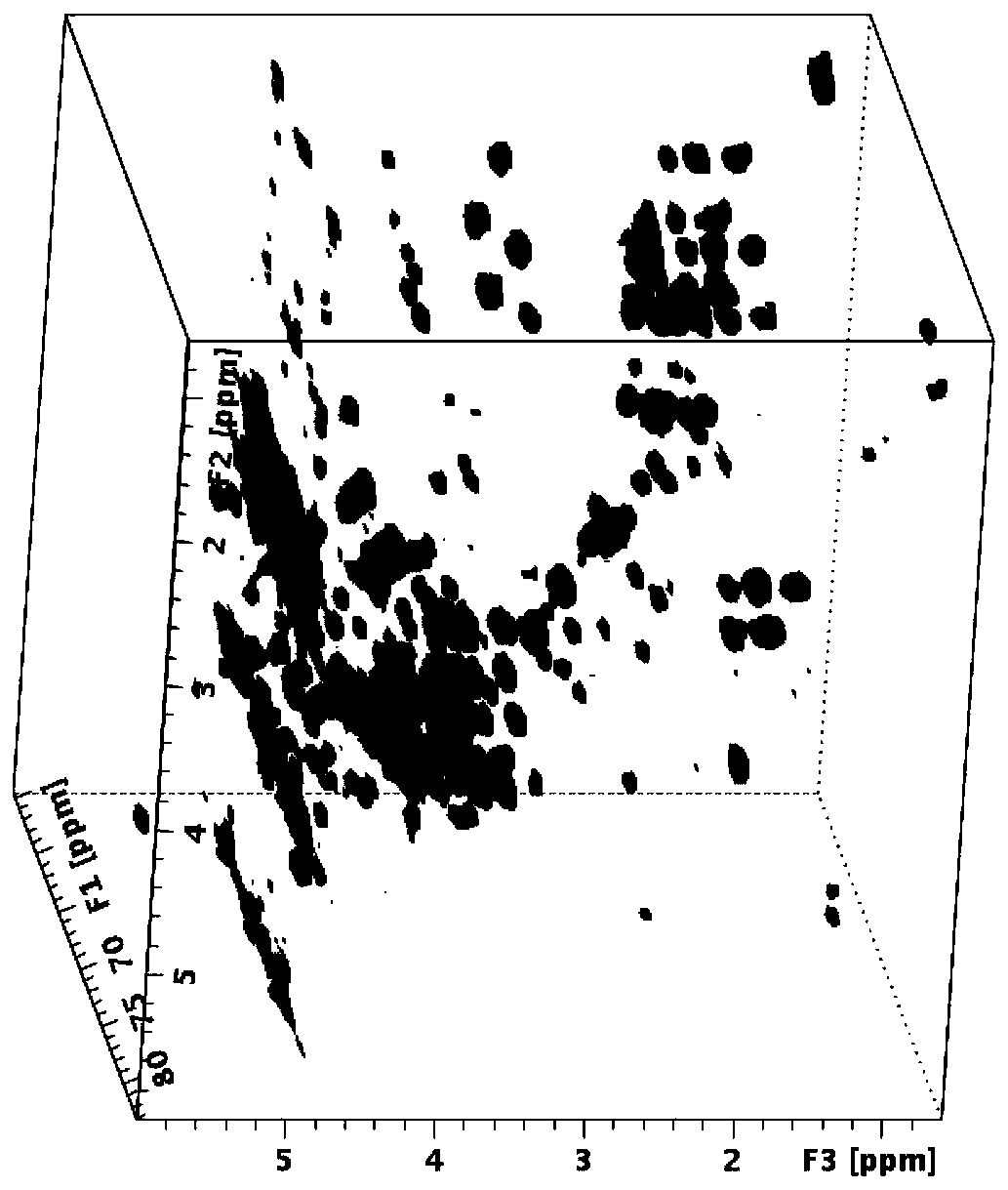
[0045] 4) Perform three-dimensional Fourier transform on the collected time domain signal to obtain three-dimensional HSQC-TOCSY nuclear magnetic resonance spectrum, the experimental spectrum is shown in image 3 . It can be seen from the figure that each signal is distinguished in the ...
Embodiment 3
[0046] Example 3: Taking decompression wax oil as an example to test the rationality and universality of a method for obtaining a highly sensitive three-dimensional nuclear magnetic resonance spectrum described in the present invention.
[0047] 1) The configuration of the sample to be tested and the optimization of the hydrogen spectrum width are the same as in Example 1.
[0048] 2) The method for optimizing the spectrum width of carbon is the same as in Example 1. After optimization, the spectrum width of carbon is 45 ppm, and that of hydrogen is 11 ppm.
[0049] 3) Setting the present invention to obtain a highly sensitive three-dimensional HSQC-TOCSY experiment is the same as in Example 1.
[0050] 4) Perform three-dimensional Fourier transform on the collected time domain signal to obtain three-dimensional HSQC-TOCSY nuclear magnetic resonance spectrum, the experimental spectrum is shown in Figure 4 . It can be seen from the figure that each signal is distinguished in...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com