Reference gene for detecting expression level of nucleic acid in exosome and application thereof
An internal reference gene and expression level technology, which is applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of poor data repeatability and instability, and improve the amplification effect and detection efficiency , high accuracy and high stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
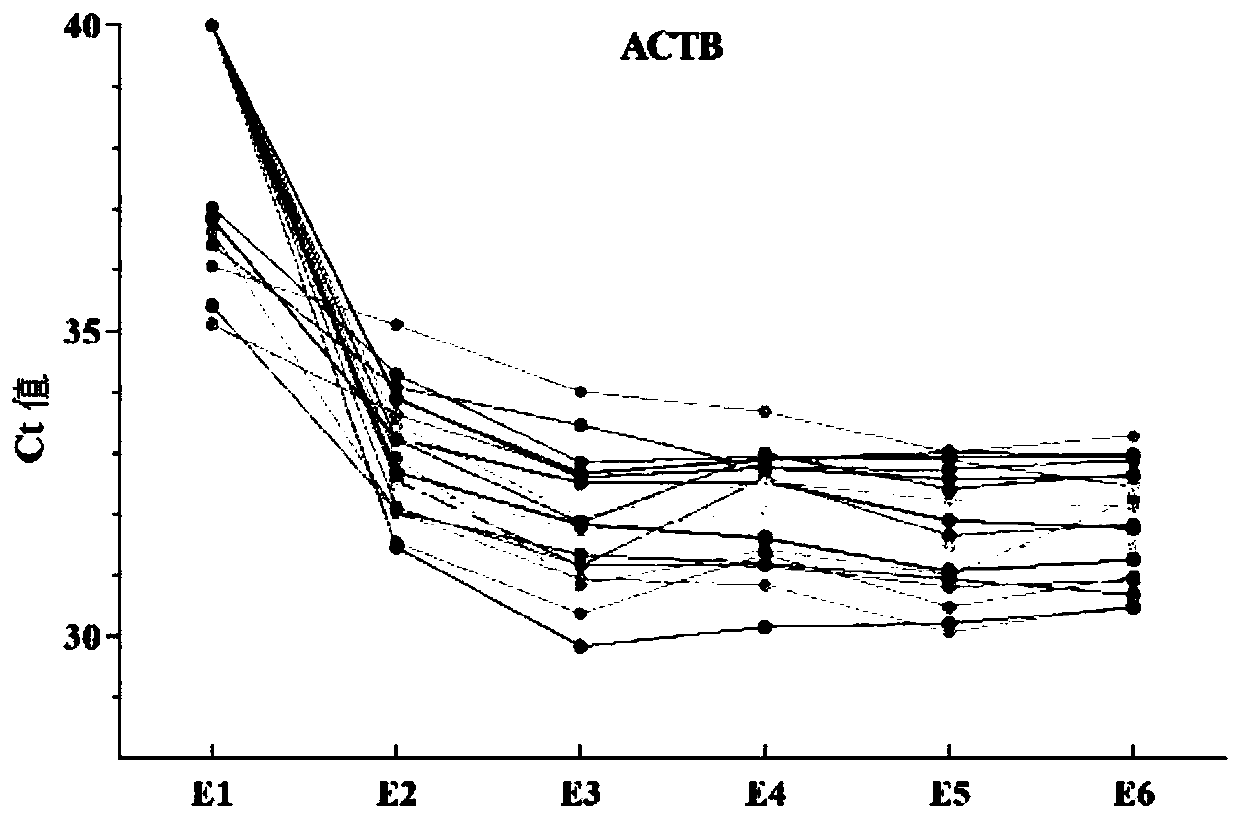
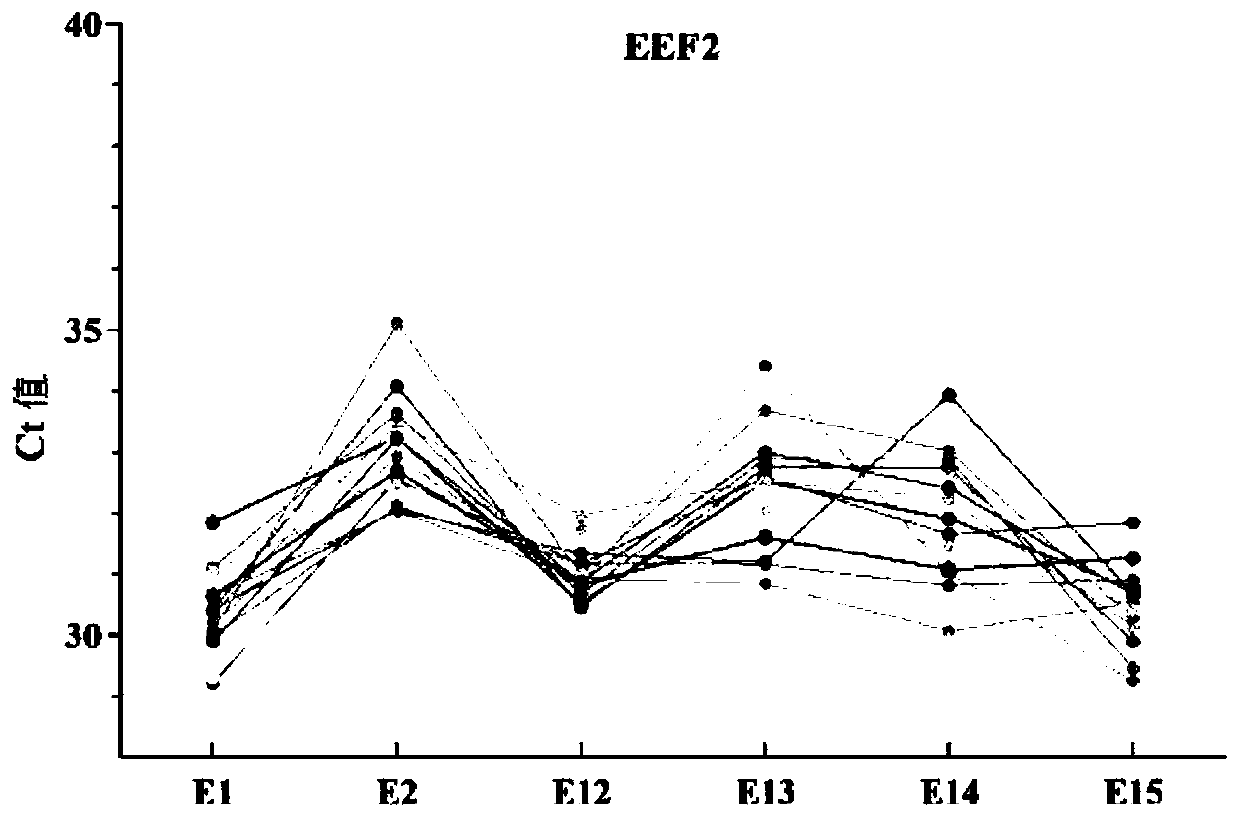
[0050] Through the following experiments, the inventors optimized the different exons of the gene according to the CT value stability of the different exons of the internal reference gene, and obtained the preferred exons of each internal reference gene.
[0051] Design primers for each gene exon: ACTB covers exons 1, 2, 3, 4, 5, and 6; EEF2 covers exons 1, 2, 12, 13, 14, and 15; MSN covers exons 1, 2 , 3, 4, 12 and 13 exons; TLN1 covers exons 1, 2, 3, 27, 56 and 57.
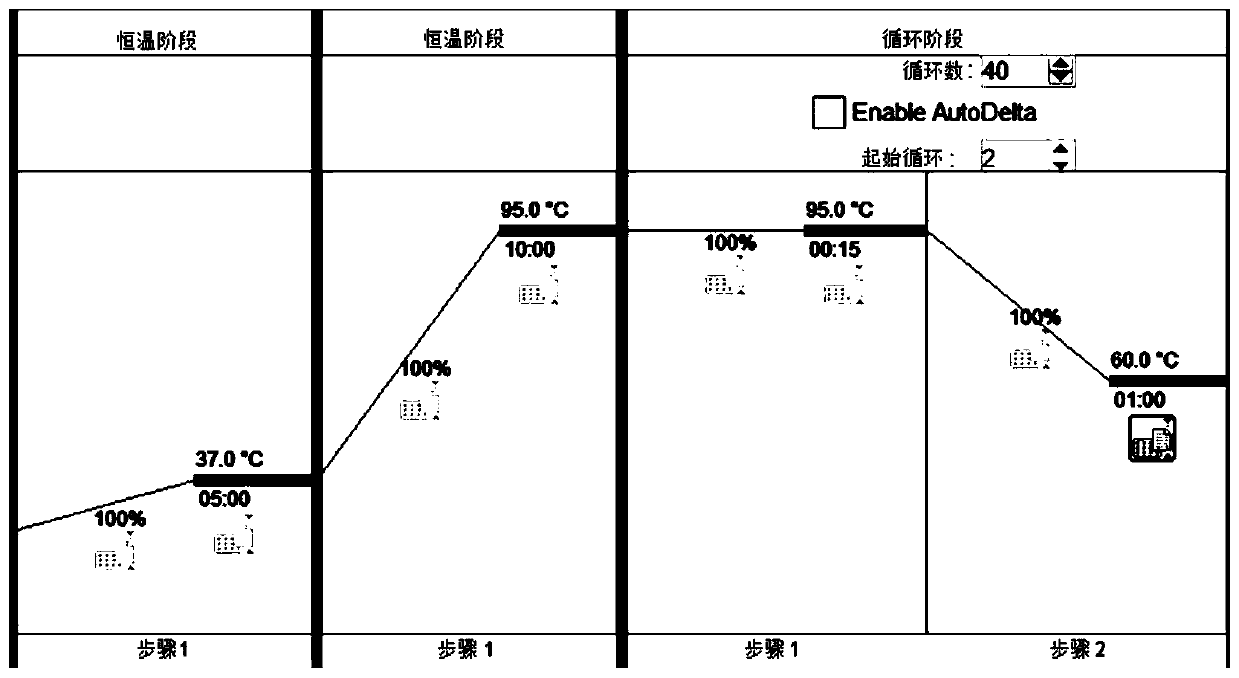
[0052] Collect blood samples from 15 clinically diagnosed patients with different diseases, and process the incoming samples as follows: use an exosome extraction kit (QIAGEN: exoEasy Maxi Kit) to extract the total exosomes in the sample serum come out. Subsequently, the extracted exosomes were extracted with a ribonucleic acid extraction kit (QIAGEN company: miRNeasy MicroKit) to extract all the ribonucleic acids in the exosomes. The extracted ribonucleic acid was used reverse transcription kit (Takara compan...
Embodiment 2
[0056] In order to further optimize the formula model of the internal reference gene combination, the inventors conducted the following research experiments to compare the differences of the internal reference model when different exon combinations were selected. Among them, the internal reference model is CONCT=0.7×ACTB CT +0.1×EEF2 CT +0.1×MSN CT +0.1×TLN1 CT .
[0057] Combination 1: ACTB selects exon 6, EEF2 selects exon 15, MSN selects exon 4, and TLN1 selects exon 57.
[0058] Combination 2: ACTB selects exon 5, EEF2 selects exon 12, MSN selects exon 12, and TLN1 selects exon 27.
[0059] Combination 3: ACTB selects exon 4, EEF2 selects exon 1, MSN selects exon 1, and TLN1 selects exon 1.
[0060] Using the blood samples of 10 clinically diagnosed patients with different diseases, compare the experimental results of the internal reference model under the above three combinations (combination 1, combination 2 and combination 3), such as Image 6 shown. For specific...
Embodiment 3
[0064] The inventors compared the expression level of the target gene in exosomes by adding an exogenous reference gene (a synthetic GH1 (growth hormone 1) gene mRNA whose sequence is shown in SEQ ID NO: 22) and the CONCT model (combination 1) strengths and weaknesses in the analysis.
[0065] The PTEN gene with high expression in prostate cancer cells was selected as the verification target. The recognized GAPDH was selected as the internal reference gene for cellular RNA, and the CONCT model and exogenous spike-in were used as internal references for the detection of PTEN expression in exosomes.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com