Four sets of oligonucleotide sequences for recognition and identification of vibrio anguillarum, and screening method of oligonucleotide sequences
An oligonucleotide and screening method technology, which is applied in the field of identification and detection of Vibrio eel, can solve the problems of increasing screening links and labor intensity, inability to amplify by PCR, and increasing screening workload, etc., so as to avoid aptamers. The risk of sequence loss or spatial structure being destroyed, the effect of reducing workload, and shortening the number of screening rounds
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
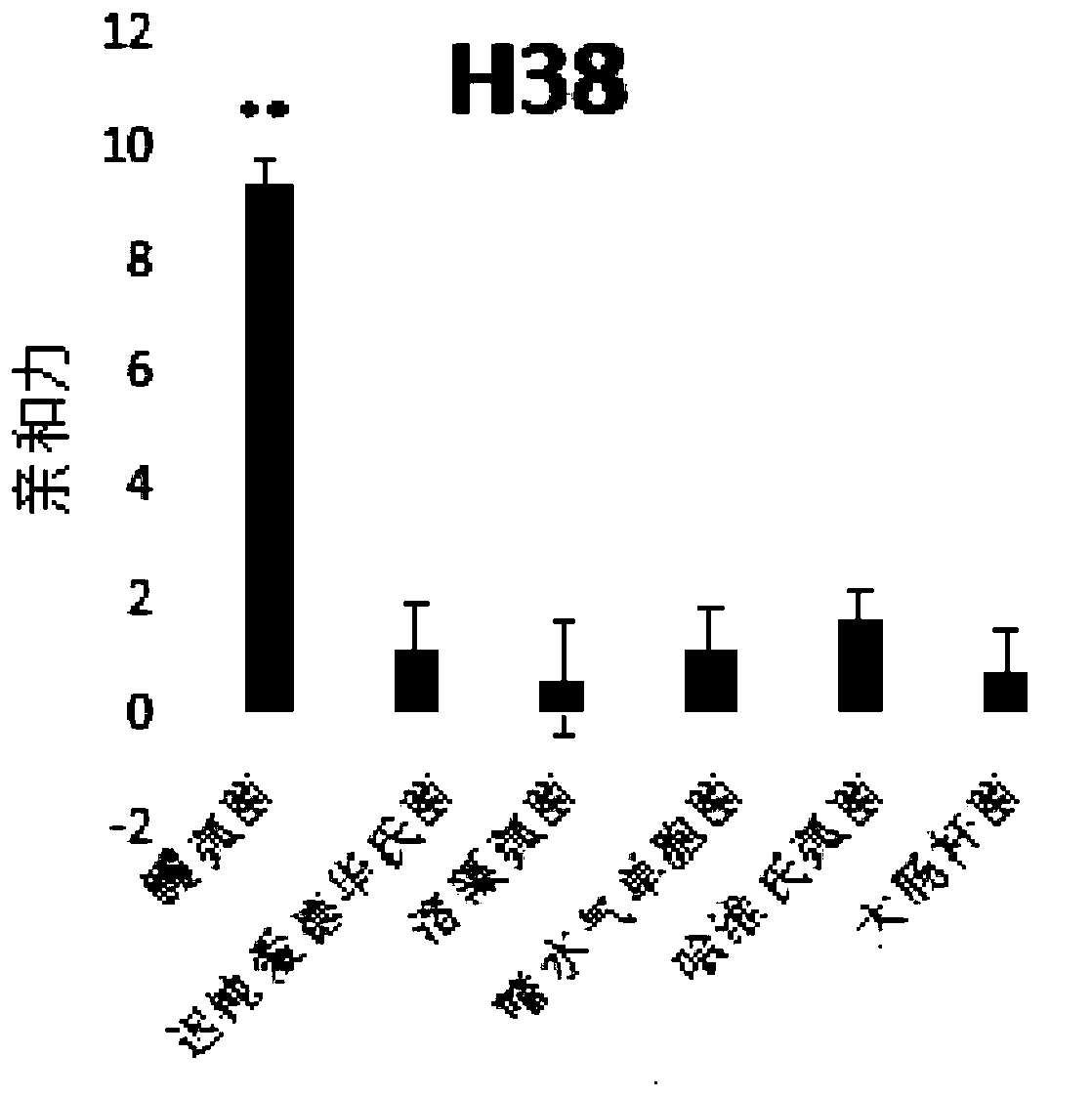
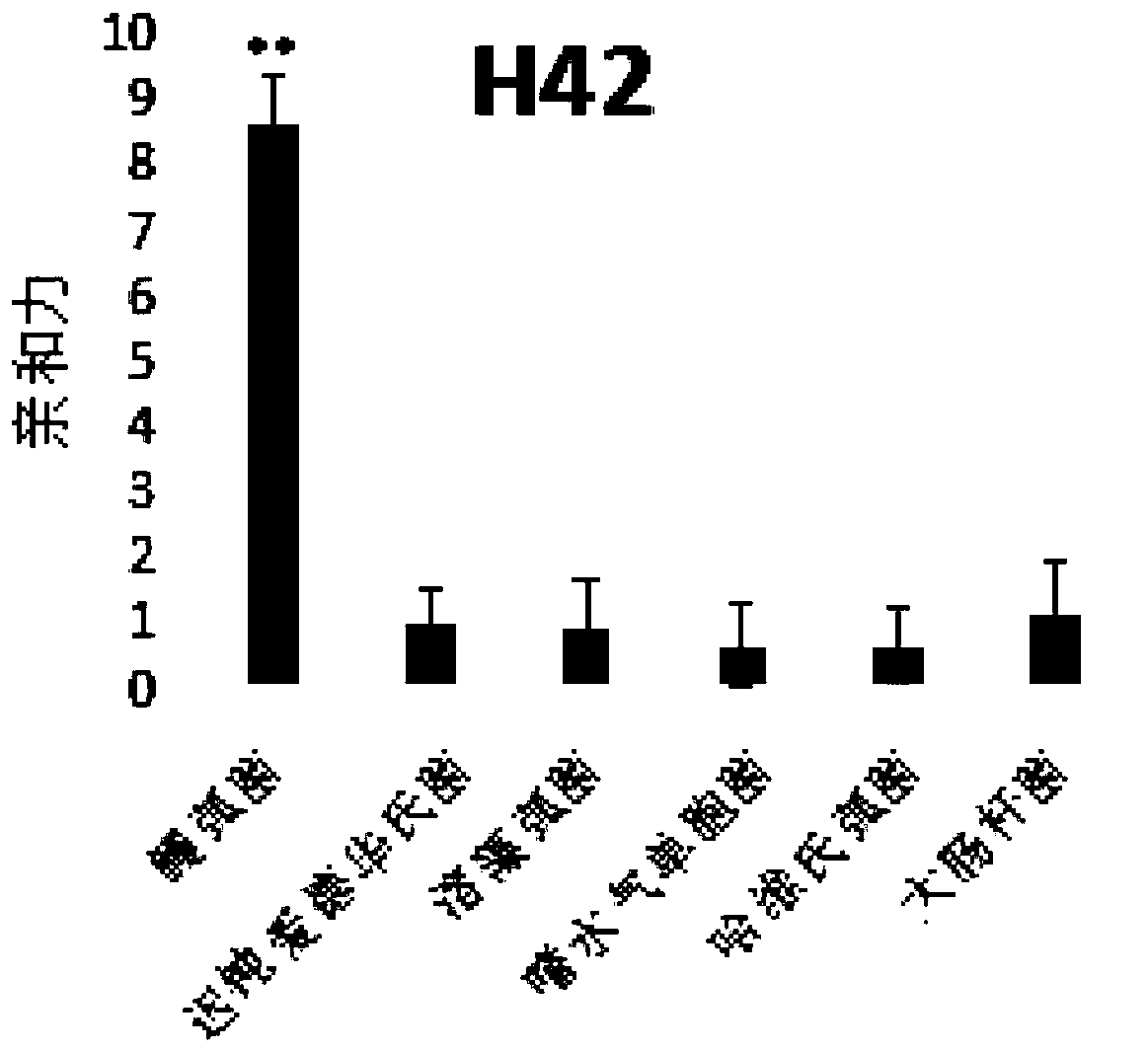
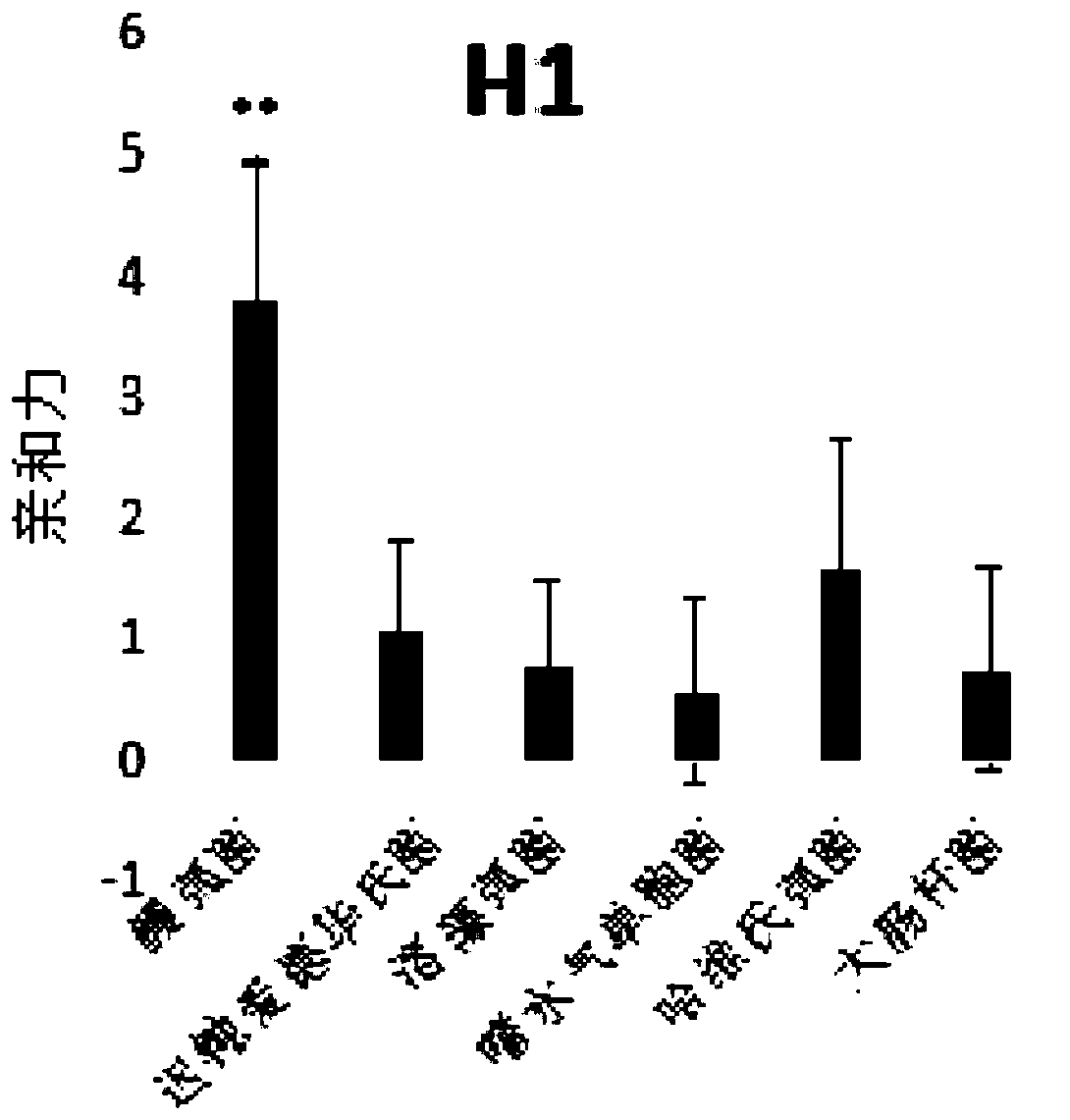
[0096] According to the above screening method, the applicant randomly synthesized the fixed sequence at both ends as 5'-TCA GTC GCT TCG CCG TCTCCT TC--the middle sequence--GCA CAA GAG GGA GAC CCC AGA GGG-3', the middle sequence is 35 bases The length of the library was repeated for 5 rounds of screening, that is, n=5. By comparing the results of high-throughput sequencing, sequences with higher frequencies were obtained. The results are as follows:
[0097]
[0098] Applicants are divided into four groups based on different sequence lengths.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com