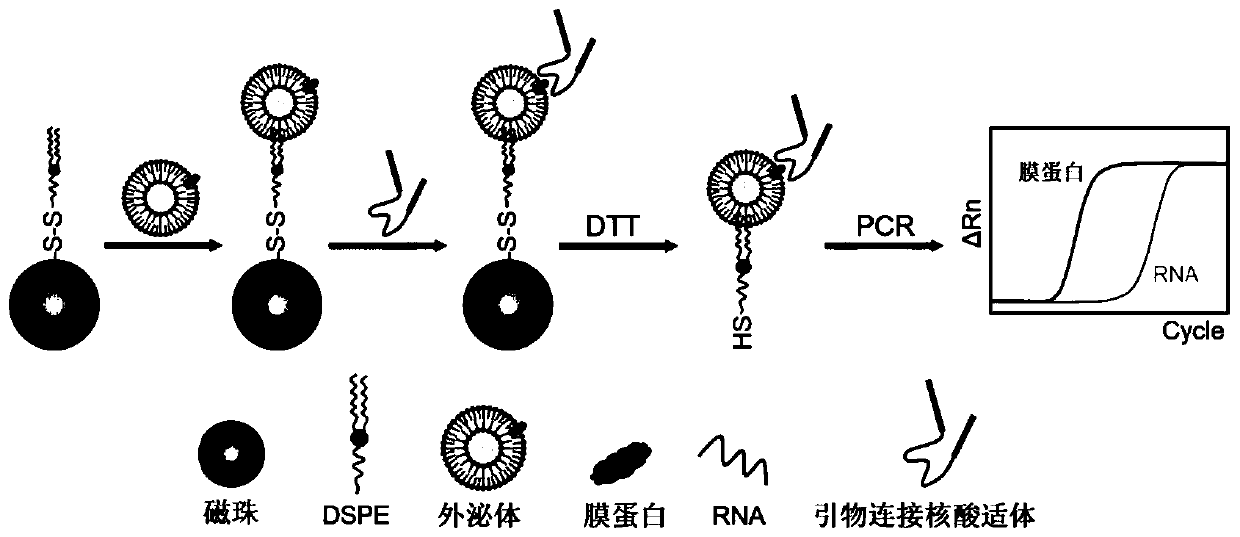
Method for jointly characterizing exosome membrane markers and RNA based on aptamer immune PCR
A nucleic acid aptamer and exosome technology, which is applied in biochemical equipment and methods, microbial measurement/inspection, animal cells, etc., can solve the problem of simultaneous detection of exosome protein and RNA, and save test samples , good specificity and stable chemical properties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1: Establishment of an aptamer-based immuno-PCR detection method
[0053] Step 1: Synthesis of DSPE-S-S-MBs;
[0054] Step 1.1: Wash 1 mL of carboxy-modified magnetic beads 3 times with PBS, and finally resuspend in 1 mL of PBS;
[0055] Step 1.2: Add 15 mg of cystamine hydrochloride to step 1.1 and mix well, then add 10 mg of EDC ((1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride) and 10 mg of hydroxysuccinimide (N-Hydroxysuccinimide, NHS ), stirring and reacting at room temperature for 5h;
[0056] Step 1.3: Separate the mixed solution in step 1.2 under an external magnetic field, repeat washing 3 times, and finally resuspend in 1mL PBS;
[0057] Step 1.4: Add 10 mg NHS and DSPE co-modified polyethylene glycol (NHS-PEG-DSPE) to step 1.3, mix well, and react at room temperature for 16 hours;
[0058] Step 1.5: Separate the mixed solution in step 1.4 under an external magnetic field, repeat washing 5 times, and finally resuspend in 1mL PBS.
[005...
Embodiment 2
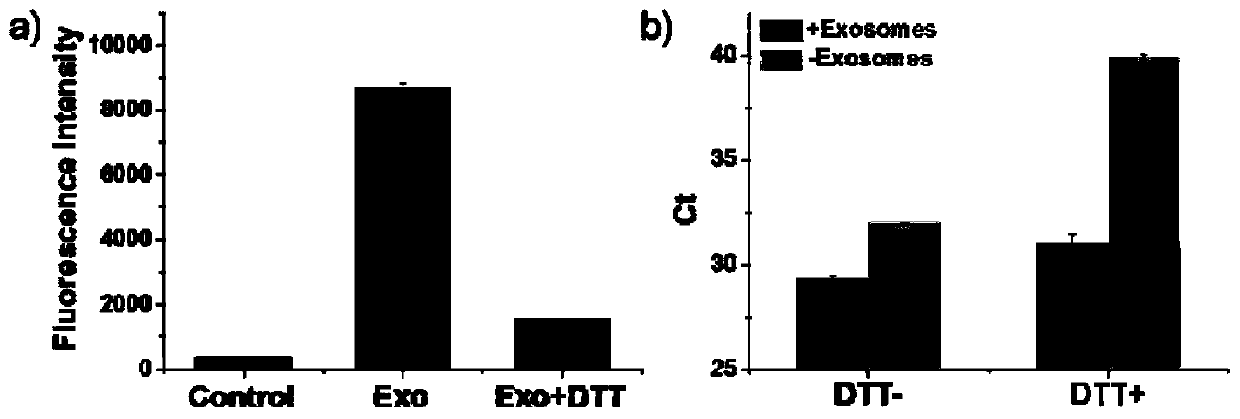
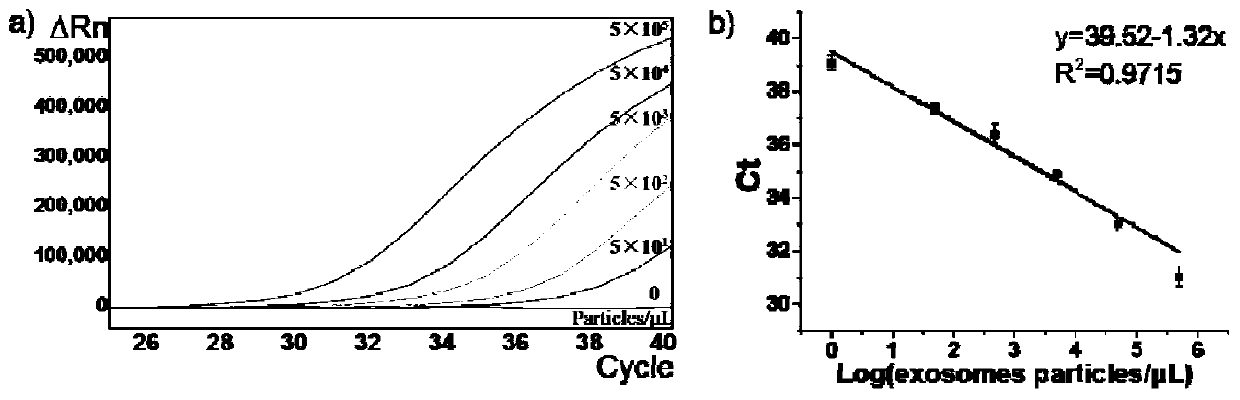
[0089] Example 2: Characterization of Exosome Membrane Marker CD63 Protein by Fluorescent Quantitative PCR for Nucleic Aptamers
[0090] Taking exosomal CD63 protein as an example, the nucleic acid aptamer shown in SEQ ID NO:1 was used to detect different concentrations (50-5×10 5 Each / μL) of exosomes, using the nucleic acid aptamer SEQ ID NO:1 of CD63 protein to perform fluorescent quantitative PCR detection, the amplification primers include SEQ ID NO:5-6, and the probe includes SEQ ID NO:7 (5' The end is labeled FAM, the 3' end is labeled MGB). The result is as image 3 shown. It can be seen that the number of cycles and Ct values detected by fluorescence quantitative PCR of nucleic acid aptamers combined with CD63 protein are negatively correlated with the concentration of exosomes. Therefore, the detection of nucleic acid aptamers by fluorescent quantitative PCR can quantitatively detect exosomal membrane markers.
Embodiment 3
[0091] Example 3: Characterization of exosomal PD-L1 protein, exosomal RNA SLC25A6 and IDO1 in the same reaction system
[0092] The exosomal PD-L1 protein, exosomal RNA SLC25A6 and IDO1 in the plasma samples of 3 healthy people (H1-H3) and 5 lung cancer patients (P1-P5) were simultaneously characterized in the same reaction system.
[0093]In the reaction system, the nucleic acid aptamer SEQ ID NO: 2 of the PD-L1 protein is detected by fluorescent quantitative PCR, the amplification primers include SEQ ID NO: 8-9, and the probe includes SEQ ID NO: 7 (the 5' end labeled FAM , 3' end labeled MGB). Real-time quantitative PCR detection for exosomal RNA SLC25A6 and IDO1, primers for exosomal RNA IDO1 include SEQ ID NO:11-12, probes include SEQ ID NO:13 (5' end labeled VIC, 3' end labeled BHQ1 ); the primers of exosomal RNA SLC25A6 include SEQ ID NO: 14-15, and the probe includes SEQ ID NO: 16 (the 5' end is marked with Cy5, and the 3' end is marked with BHQ3).
[0094] The resul...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com