Mmlv reverse transcriptase variants
A technology of reverse transcriptase and reverse transcription, applied in the direction of transferase, enzyme, hydrolase, etc., can solve problems such as poor solution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
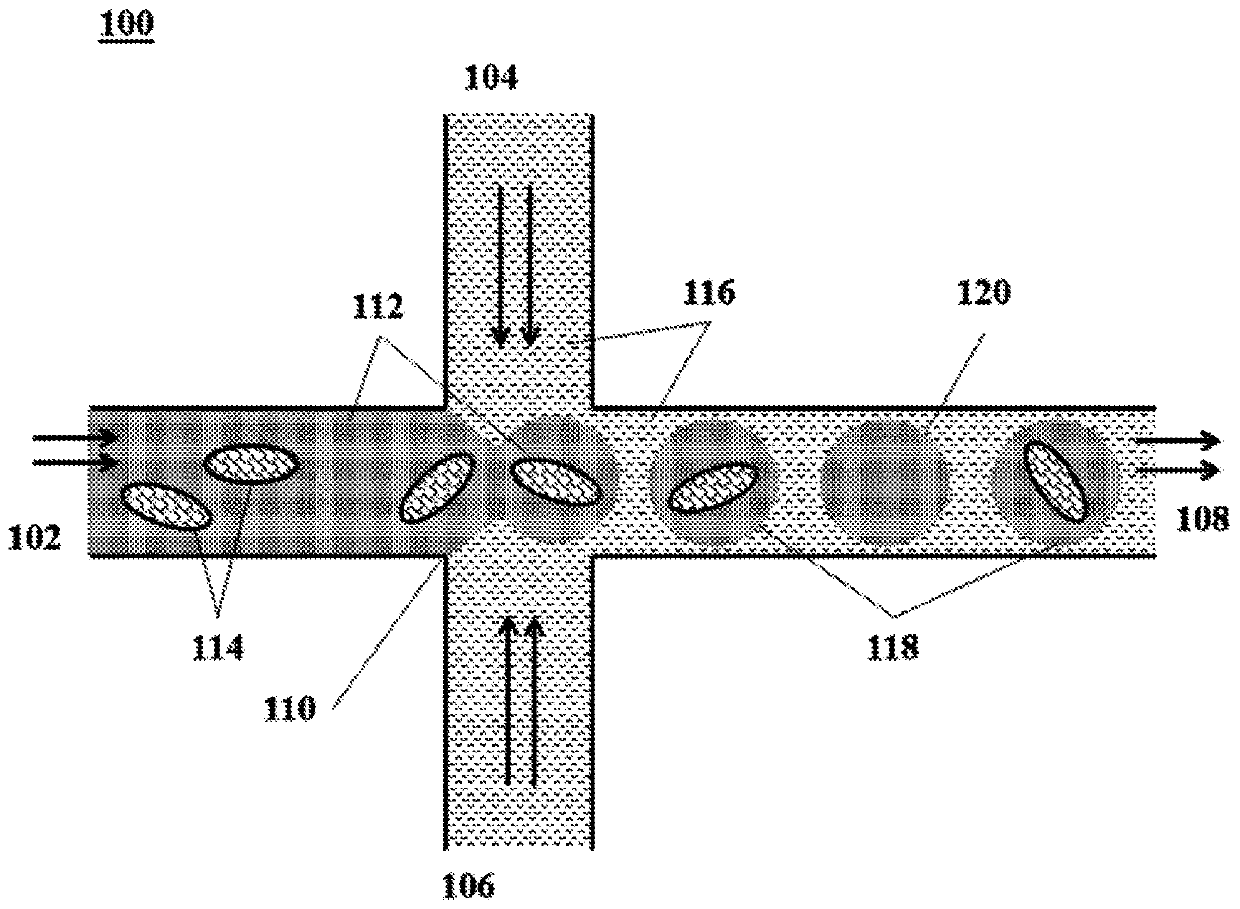
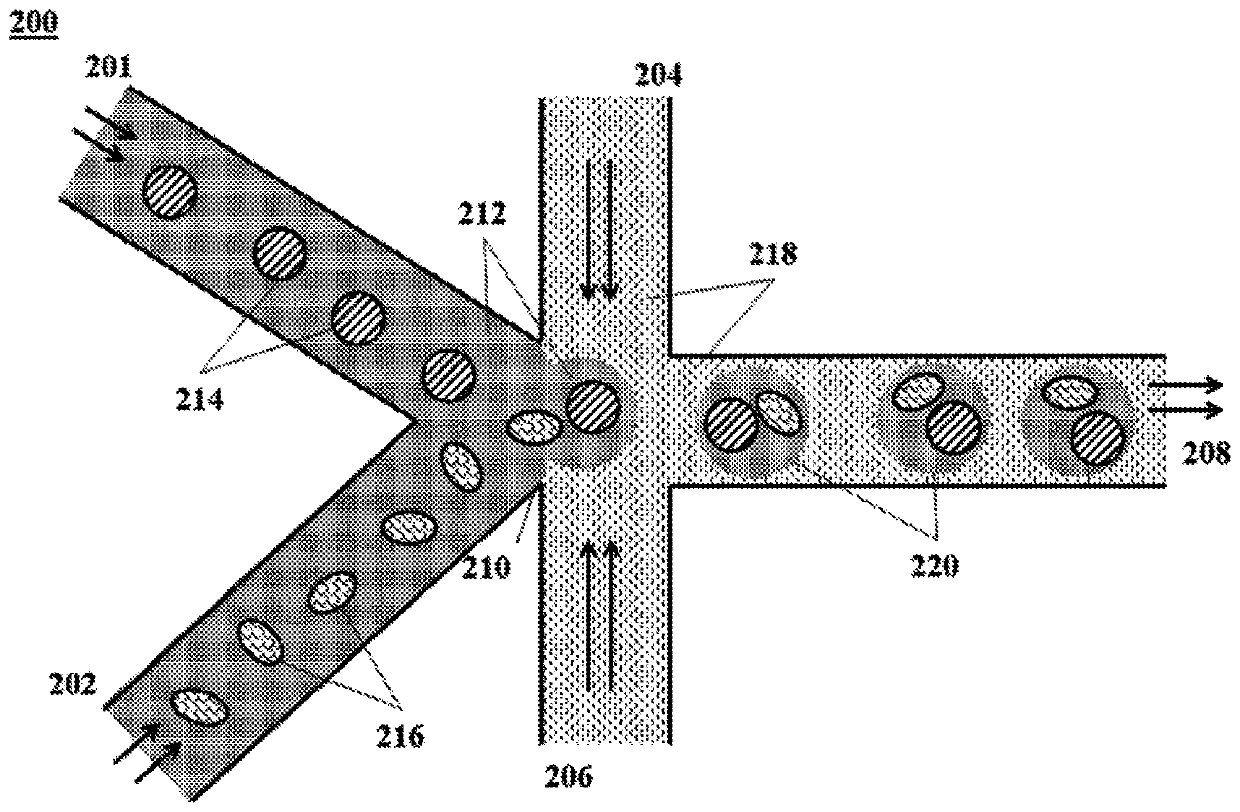
[0066] Preparation of cell-containing microcapsules can be performed in a number of ways. For example, an air knife droplet or aerosol generator can be used to dispense droplets of precursor fluid into the gelling solution to form microcapsules including single cells or small groups of cells. Likewise, membrane-based encapsulation systems can be used to produce microcapsules comprising encapsulated cells as described herein. Disclosures such as those shown in figure 1 The microfluidic system in can be readily used to encapsulate cells as described herein. In particular, and with reference to figure 1 , an aqueous fluid 112 containing (i) individual cells 114 and (ii) polymer precursor material (not shown) flows into the channel junction 110 where it is degraded by the flow of a non-aqueous fluid 116 Dispensed into droplets 118,120. In the case of an encapsulation method, the non-aqueous fluid 116 may also include an initiator (not shown) to cause polymerization and / or cros...
Embodiment 1
[0313] Example 1. Analysis of cellular RNA using emulsions
[0314] In one instance, as in Figure 9A The procedure shown in , reverse transcription with template switching and cDNA amplification (by PCR) was performed in the case of emulsion droplets. Reaction mix dispensed for reverse transcription and cDNA amplification (by PCR) consisted of 1,000 cells or 10,000 cells or 10 ng RNA, beads with barcoded oligonucleotides / 0.2% Tx-100 / 5x Kapa buffer , 2x Kapa HS HiFi Ready Mix, 4 μM Switching Oligomer and Smartscribe. When cells are present, the mixture is partitioned such that most or all droplets contain a single cell and a single bead. The cells are lysed while the barcoded oligonucleotides are released from the beads, and the poly-dT segment of the barcoded oligonucleotides hybridizes to the poly-A tail of the mRNA released from the cells, as in operation 950 . As in operation 952, the poly-dT segment is extended in a reverse transcription reaction, and as in operation 9...
Embodiment 2
[0316] Example 2. Analysis of cellular RNA using emulsions
[0317] In another example, as in Figure 9A The procedure shown in , reverse transcription with template switching and cDNA amplification (by PCR) was performed in the case of emulsion droplets. The reaction mix dispensed for reverse transcription and cDNA amplification (by PCR) included Jurkat cells, beads carrying barcoded oligonucleotides / 0.2% Triton X-100 / 5x Kapa buffer, 2xKapa HS HiFi Ready Mix , 4 μM switching oligo and Smartscribe. The mixture is partitioned such that most or all droplets contain single cells and single beads. The cells are lysed while the barcoded oligonucleotides are released from the beads, and the poly-dT segment of the barcoded oligonucleotides hybridizes to the poly-A tail of the mRNA released from the cells, as in operation 950 . As in operation 952, the poly-dT segment is extended in a reverse transcription reaction, and as in operation 954, the cDNA transcript is amplified. Therma...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com