Method for storing information in vivo by using DNA
A technology for internal storage and encoding of information, applied in recombinant DNA technology, methods based on microorganisms, biochemical equipment and methods, etc., can solve the problems of high synthesis and storage costs, difficult storage, short storage time, etc., to achieve low cost, Improved storage capacity and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
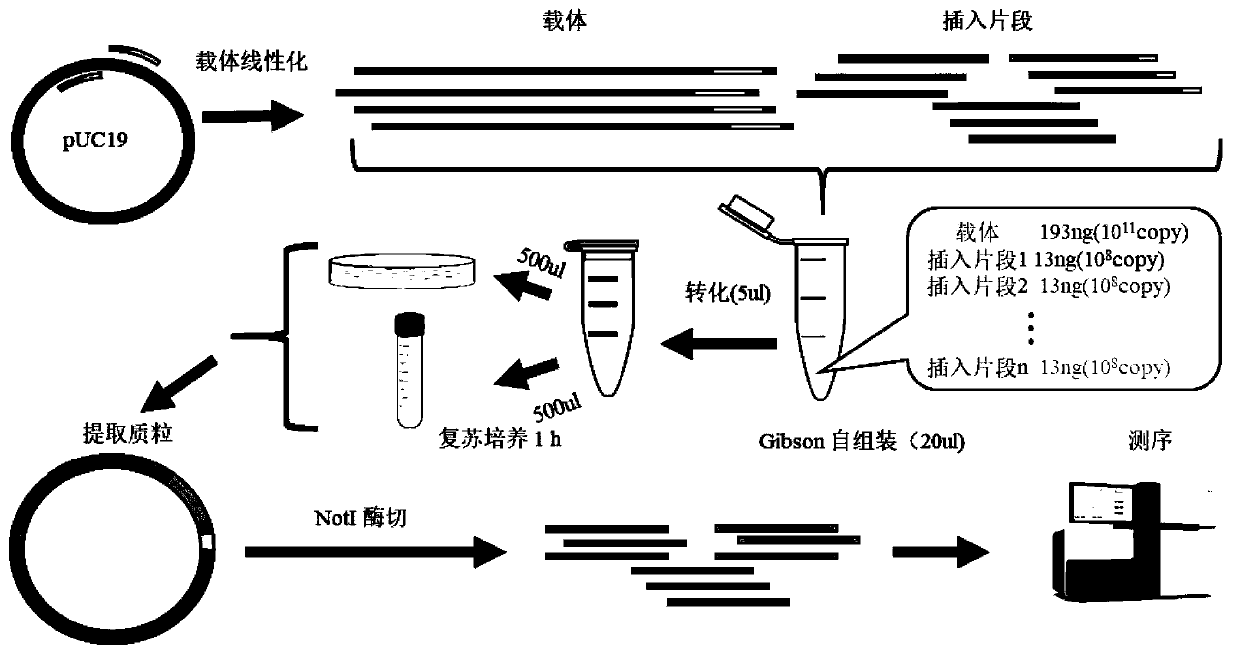
[0033] Embodiment 1, PCR preparation insert fragment and carrier
[0034] 1. Preparation of insert fragments
[0035] Q5 Master pool concentration: 14.024ng / μl
[0036] Table 1 insert
[0037] number of molecules copies per molecule Copy number corresponds to quality 12000 10 5
0.233ng
[0038] Dilute the master pool 200 times to a concentration of 0.07ng / μl.
[0039] The reaction system for PCR amplification using Q5 hot-start ultra-fidelity DNA polymerase is shown in Table 2.
[0040] Table 2 reaction system
[0041]
[0042]
[0043] The sequences of the forward and reverse primers are shown in Table 6.
[0044] The reaction program of PCR amplification is shown in Table 3.
[0045] Table 3 Reaction program
[0046]
[0047] After the PCR product is purified and recovered, use 30 μl ddH 2 O was dissolved, and the concentration of insert fragments was obtained by grayscale analysis.
[0048] 2. Preparation of carrier fragments ...
Embodiment 2
[0059] Embodiment 2, recombinant plasmid construction
[0060] 1, 3 or 5 inserts with corresponding homology arms were ligated with pUC19 vector using Gibson assembly kit, and reacted at 50°C for 60 min. The specific reaction system is shown in Table 7-9 below.
[0061] Table 7 The reaction system (pUC19-assembly-1) in which an insert fragment is connected to the pUC19 vector
[0062] components Dosage (μl) pUC19 linearized vector 2μl (200ng) insert fragment 1 1μl (13ng) Gibson Assembly Master Mix(2×) 10μl wxya 2 o
7μl
[0063] Table 8 The reaction system (pUC19-assembly-3) in which three inserts were connected to the pUC19 vector
[0064] components Dosage (μl) pUC19 linearized vector 2μl (200ng) insert fragment 1 1μl (13ng) insert fragment 2 1μl (13ng) insert fragment 3 1μl (13ng) Gibson Assembly Master Mix(2×) 10μl wxya 2 o
5μl
[0065] Table 9 The reaction syste...
Embodiment 3
[0067] Embodiment 3, recombinant plasmid transformation and cultivation
[0068] The plasmid assembled by Gibson was electrotransformed into E. coli competent cells, and the specific operations were as follows:
[0069] 1. Insert an electric cup with an electrode spacing of 0.1cm into crushed ice, and let it stand in the ice for 5 minutes to fully cool down the electric cup.
[0070] 2. Take the electroporation competent cells (Escherichia coli DH10β) stored at -70°C, the transformation efficiency is greater than 10 10 cfu / μgDNA) into the ice, after the cells have just thawed, add 5 μl of the ligated recombinant plasmid, immediately insert into the ice, and quickly transfer the cell / DNA mixture to the electric shock cup with a sterile tip in the ultra-clean bench to avoid Bubble, make sure the cells sink to the bottom of the cup, cover the cup, and keep the empty tube for later use.
[0071] 3. Start the electrorotator and set the shock parameters: 1.8kV. Put the electric c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com