Nonenzymatic biosensor for detecting activity of uracil-DNA glycosylase
A biosensor and glycosylase technology, applied in the field of biosensors, can solve the problems of complex operation, high cost, low specificity and sensitivity, etc., and achieve the effect of good repeatability, simple operation and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
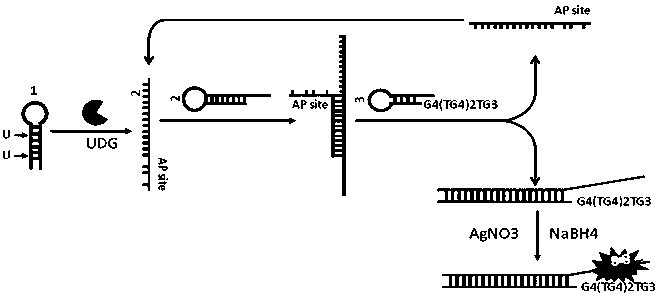
[0035] Example 1 Preparation of DNA Templated Silver Nanoclusters
[0036] Formulating AgNO 3 The concentration is 100 μM, the volume is 1 mL, and stored in the dark at 4°C. Prepare NaBH 4Concentration 100 μM, volume 1 mL, NaBH 4 It is ready-to-use and prepared with ice water at 0°C.
[0037] The solution after catalytic hairpin self-assembly contains 2 strands (5 μL, 10 μM), so add the prepared AgNO 3 (100μM) solution 3.0μL, after shaking for 30s, place it in a dark place at 4°C for 20min, then take it out and add freshly prepared NaBH 4 (100 μM) solution 3.0 μL, after shaking for 30 s, place at 4 °C in the dark for 3 h.
Embodiment 2
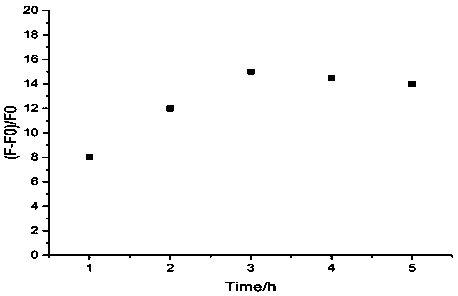
[0038] Example 2 Fluorescence intensity changes with incubation time.
[0039] (1) Mix 2 μL of chain 1 (5 μM), 1 μL of UDG (100 U / mL) and 10 μL of UDG-10×Reaction Buffer, and react at 37°C for 1 hour. The UDG-10×Reaction Buffer: contains 20 mM Tris-HCl (pH=8.2, 25° C.), 10 mM EDTA, and 100 mM NaCl.
[0040] (2) Heat the solution in step (1) at 90°C to inactivate UDG enzyme, then cool it down to 20°C for later use.
[0041] (3) Add 10 μL of 2-strand (5 μM) and 10 μL of 3-strand (5 μM) to the solution in step (2) to carry out catalytic hairpin self-assembly reaction at 20° C. for 2 h.
[0042] (4) Add AgNO to the solution in step (3) 3 (100μM) solution 3.0μL, after shaking for 30s, place it in a dark place at 4°C for 20min, then take it out and add freshly prepared NaBH 4 (100 μM) solution 3.0 μL, shake for 30 seconds, and add 20 mM PBS buffer to make the final reaction volume 100 μL, incubate at 4°C in the dark, and the incubation time is 1h, 2h, 3h, 4h, 5h respectively. Fl...
Embodiment 3
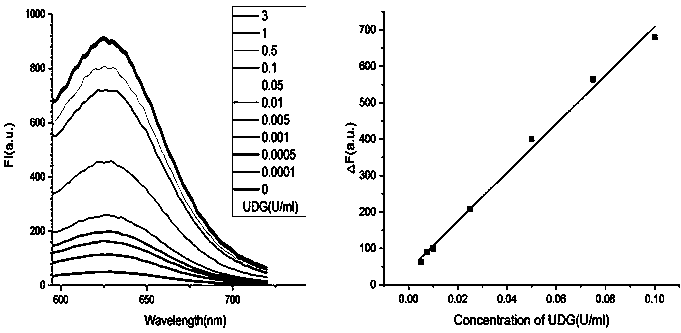
[0044] Example 3 Activity detection of UDG
[0045] (1) Mix 2 μL of chain 1 (5 μM) with different concentrations of UDG (the final concentration of UDG is 0 U / mL, 0.0001 U / mL, 0.0005 U / mL, 0.001 U / mL, 0.005 U / mL, 0.01U / mL, 0.05U / mL, 0.1U / mL, 0.5U / mL, 1U / mL, 3U / mL) and 10μL of UDG-10×Reaction Buffer were mixed and reacted at 37°C for 1h. The final volume of the reaction was 100 μL.
[0046] (2) Heat the solution in step (1) at 90°C to inactivate UDG enzyme, then cool it down to 20°C for later use.
[0047] (3) Add 10 μL of 2-strand (5 μM) and 10 μL of 3-strand (5 μM) to the solution in step (2) to carry out catalytic hairpin self-assembly reaction at 20°C for 2 h.
[0048] (4) Add AgNO to the solution in step (3) 3 (100μM) solution 3.0μL, after shaking for 30s, place it in a dark place at 4°C for 20min, then take it out and add freshly prepared NaBH 4 (100 μM) solution 3.0 μL, shake for 30 seconds, and add 20 mM PBS buffer to make the final reaction volume 100 μL, incubate ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com