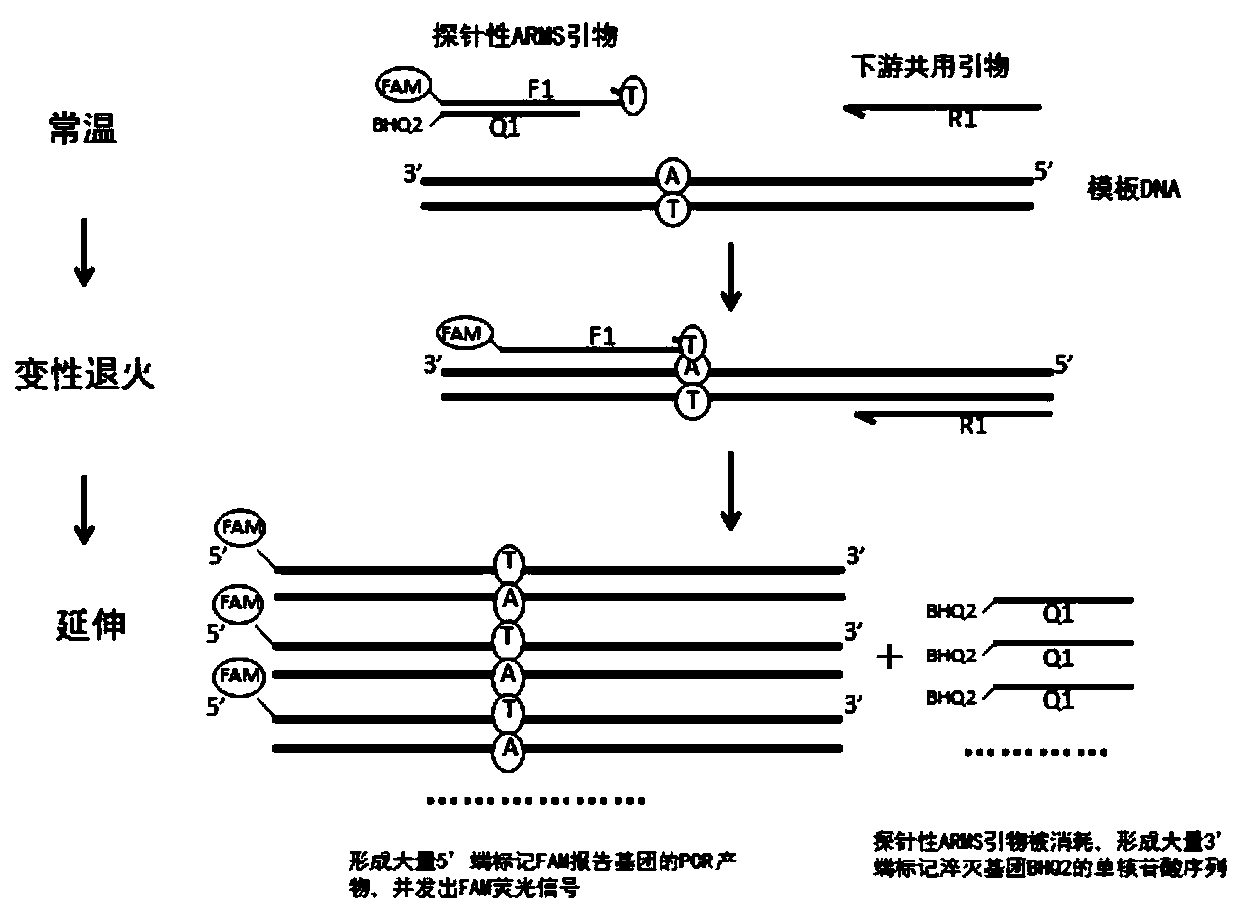
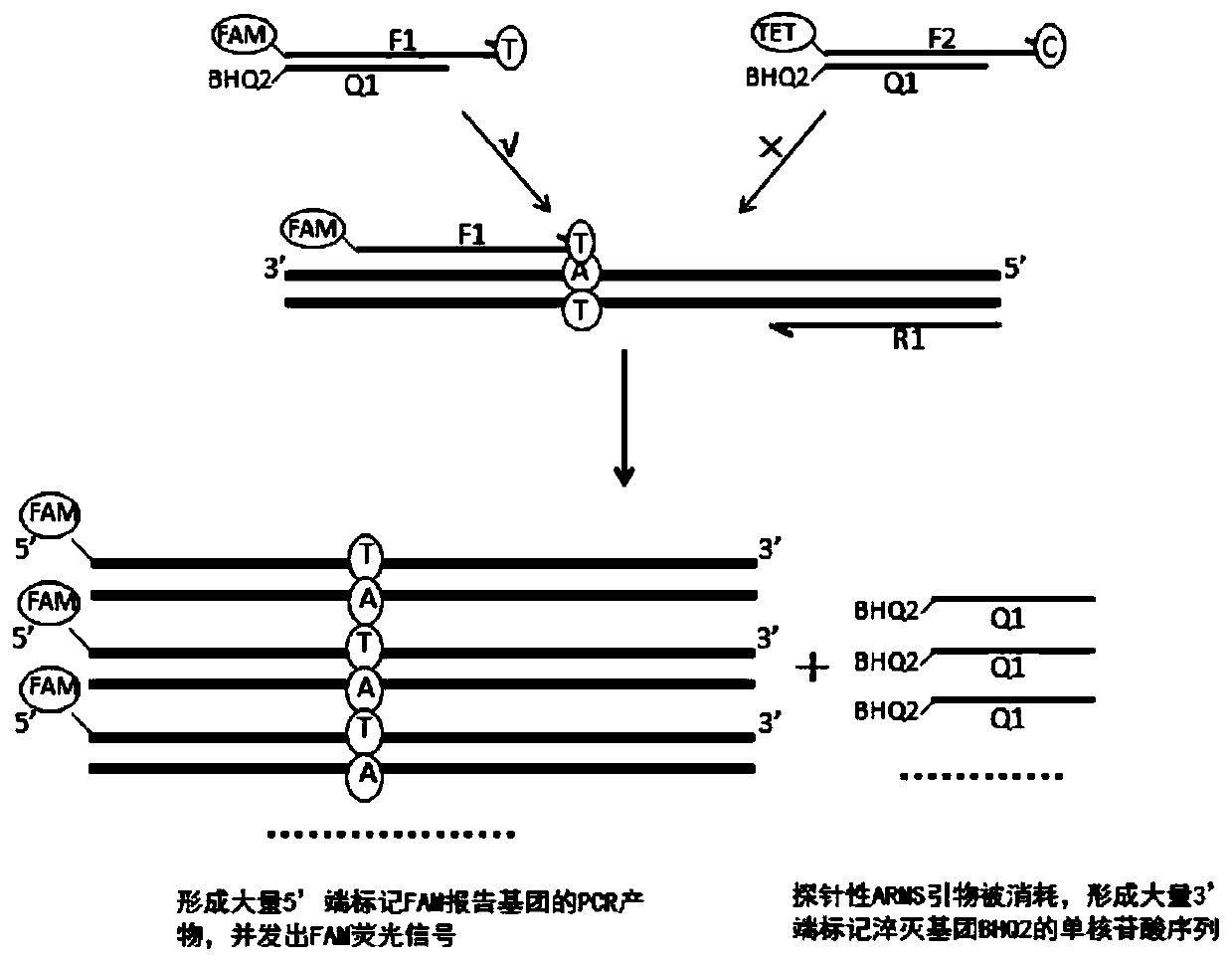
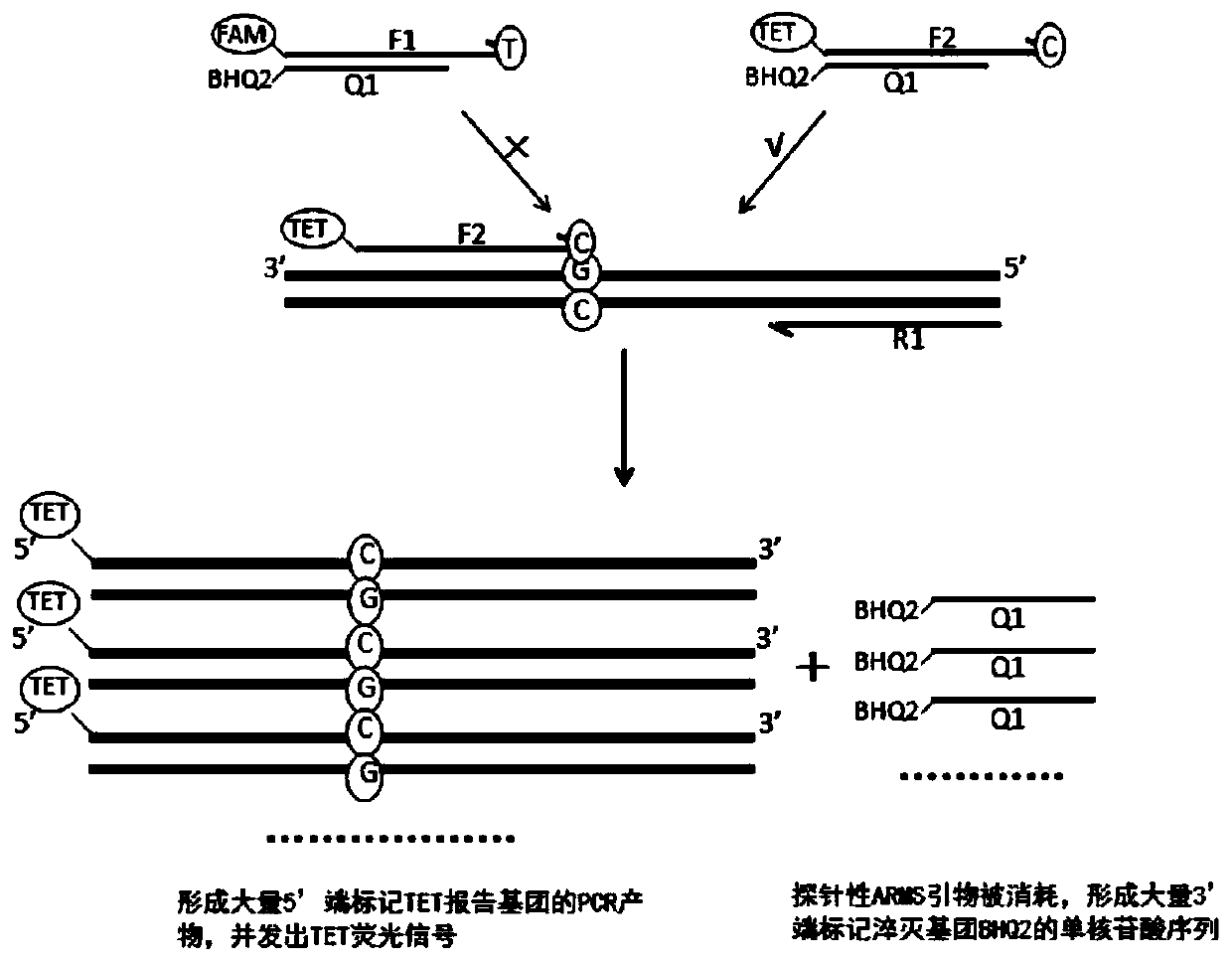
Primer group, method and kit for detecting SLCO1B1 and APOE gene polymorphism based on shared primer probe
A gene polymorphism and primer set technology, which is applied in the field of PCR detection of gene polymorphism, can solve the problems of increased adverse reactions of statins, decreased OATP1B1 uptake ability, and increased blood concentration of statins.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0149] Example 1 Design optimization of primers, optimization screening of markers
[0150] 1. The design and screening of wild-type and mutant-type probe ARMS primers, the specific steps are as follows:
[0151] 1) Preliminarily design a series of ARMS primers for SLCO1B1*5, SLCO1B1*1b, APOE2, APOE4 wild-type and mutant sites, with a Tm value of 58-62°C, and the sequences are shown in Table 1:
[0152] Table 1. Preliminary design of SLCO1B1*5, SLCO1B1*1b, APOE2, APOE4 wild-type and mutant ARMS primers
[0153]
[0154]
[0155]
[0156]
[0157] 2) SLCO1B1*5 wild-type ARMS primers, downstream primers, and SYBR-premix buffer to prepare PCR reaction solutions, respectively detect 5000 copy / μl of SLCO1B1*5 wild-type plasmids and mutant plasmids; SLCO1B1*5 mutant ARMS primers, downstream primers , SYBR-premix buffer to prepare PCR reaction solution, respectively detect 5000copy / μl SLCO1B1*5 wild-type plasmid and mutant plasmid; SLCO1B1*1b wild-type ARMS primer, downs...
Embodiment 2
[0199] Example 2 Primer preparation and kit assembly
[0200] 1. Synthesis and fluorescent labeling of probe-based ARMS primers, quenching probes, and downstream primers in this application
[0201] The probing ARMS primers, quenching probes, downstream primers and markers screened were optimized according to Example 1, and primer synthesis and fluorescent labeling were carried out. SLCO1B1*5 mutant type probe ARMS primer 5′-end labeled TET, SLCO1B1*5 quencher probe 3′-end labeled BHQ2, SLCO1B1*5 downstream primer; SLCO1B1*1b wild-type probe ARMS primer 5′-end labeled ROX , SLCO1B1*1b mutant type probe ARMS primer 5′-end labeled CY5, SLCO1B1*1b quencher probe 3′-end labeled BHQ2, SLCO1B1*1b downstream primer; APOE2 wild-type probe-type ARMS primer 5′-end labeled FAM, APOE2 mutant type probe ARMS primer 5′-end labeled TET, APOE2 quencher probe 3′-end labeled BHQ2, APOE2 downstream primer; APOE4 wild-type probe ARMS primer 5′-end labeled ROX, APOE4 mutant type probe ARMS The 5...
Embodiment 3
[0251] Embodiment 3 detection sensitivity
[0252] Use the kit a based on shared primer probes to detect human SLCO1B1 and APOE gene polymorphisms prepared in Example 2 and the detection kit b based on ARMS primers+TaqMan probes to detect SLCO1B1 and APOE gene polymorphisms, based on MGB probes Detect SLCO1B1 and APOE gene polymorphism kit c comparison test, the specific comparison test steps are as follows:
[0253] Step 1. Prepare sensitivity templates, including SLCO1B1*5 homozygous wild type 2ng / μl, 1ng / μl, 0.5ng / μl, 0.2ng / μl, SLCO1B1*5 homozygous mutant 2ng / μl, 1ng / μl, 0.5ng / μl, 0.2ng / μl, SLCO1B1*5 heterozygous 2ng / μl, 1ng / μl, 0.5ng / μl, 0.2ng / μl; SLCO1B1*1b homozygous wild type 2ng / μl, 1ng / μl, 0.5ng / μl μl, 0.2ng / μl, SLCO1B1*1b homozygous mutant 2ng / μl, 1ng / μl, 0.5ng / μl, 0.2ng / μl, SLCO1B1*1b heterozygous 2ng / μl, 1ng / μl, 0.5ng / μl , 0.2ng / μl; APOE2 homozygous wild type 2ng / μl, 1ng / μl, 0.5ng / μl, 0.2ng / μl, APOE2 homozygous mutant 2ng / μl, 1ng / μl, 0.5ng / μl, 0.2ng / μl μl, APOE...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com