Method for preparing S-3-dimethylamino-1-(2-thienyl)-1-propanol through biocatalysis
A dimethylamino, S-3- technology, applied in the field of biocatalytic preparation of S-3-dimethylamino-1--1-propanol, can solve the problems of increasing operation difficulty and cost, and achieve production cost reduction effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Through the method of whole gene synthesis, the secondary structure and codon bias of the gene are adjusted to achieve high expression in Escherichia coli.
[0031] Use Primer Premier (http: / / primer3.ut.ee / ) and OPTIMIZER (http: / / genomes.urv.es / OPTIMIZER / ) to design, and ensure that the Tm difference is controlled within 3°C, and the primer length is controlled within 60base , and the obtained primers were dissolved in double-distilled water, and added to the following reaction system, so that the final concentration of each primer was 30 nM, and the final concentration of the first and last primers was 0.6 μM.
[0032]
[0033]
[0034] The prepared PCR reaction system was placed in the Bioer XP cycler gene amplification instrument, and the amplification was carried out according to the following procedures: 98°C for 30s, 55°C for 45s, 72°C for 120s, 35x.
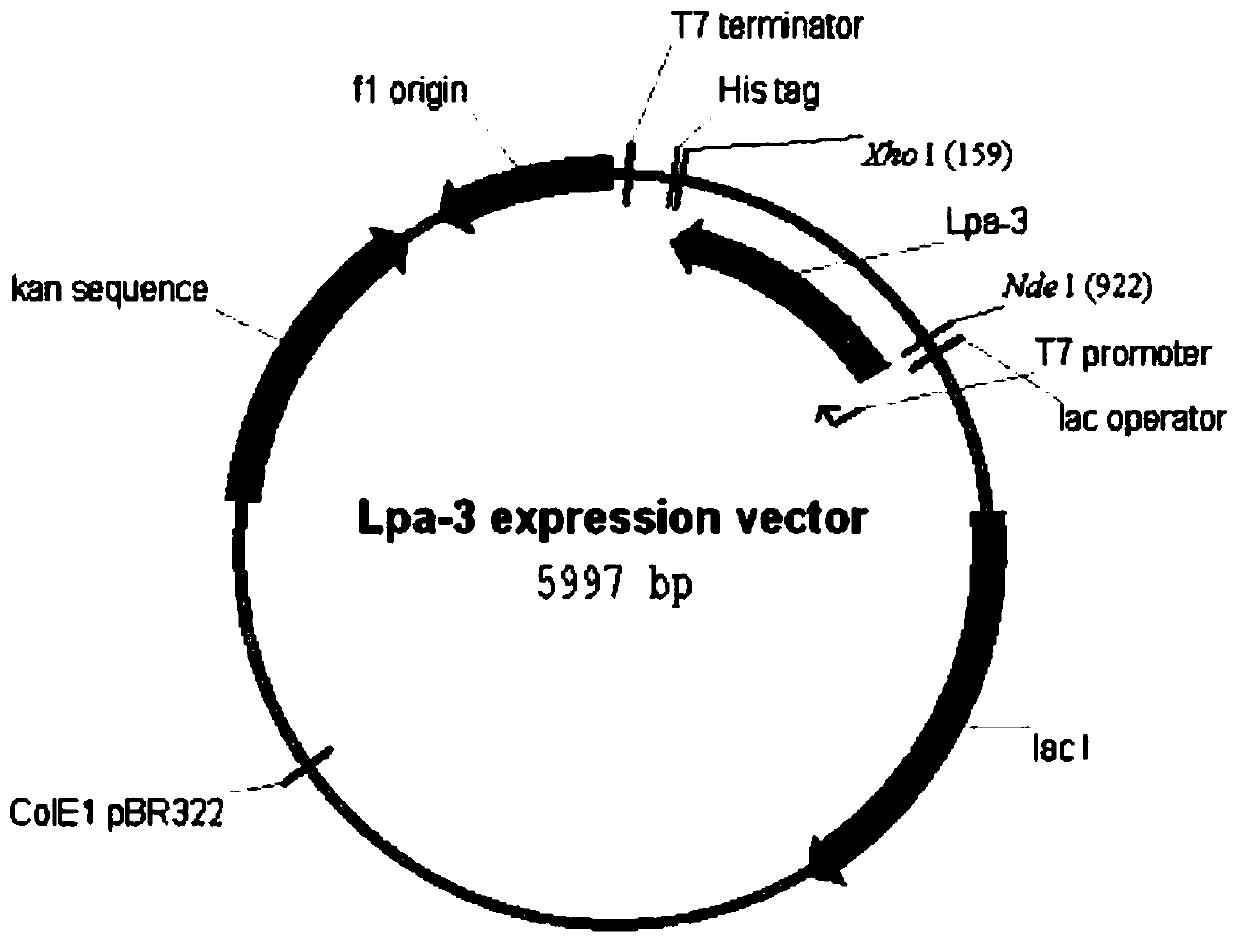
[0035] The DNA fragment obtained by PCR was gel-cut and purified, and cloned into the NdeI / XhoI site of pET...
Embodiment 2
[0038] Through the method of whole gene synthesis, the secondary structure and codon bias of the gene are adjusted to achieve high expression in Escherichia coli.
[0039] Use Primer Premier (http: / / primer3.ut.ee / ) and OPTIMIZER (http: / / genomes.urv.es / OPTIMIZER / ) to design, and ensure that the Tm difference is controlled within 3°C, and the primer length is controlled within 60base , and the obtained primers were dissolved in double-distilled water, and added to the following reaction system, so that the final concentration of each primer was 30 nM, and the final concentration of the first and last primers was 0.6 μM.
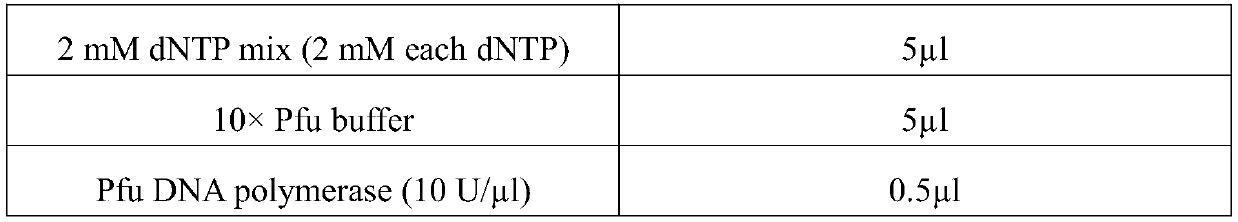
[0040] 2mM dNT Pmix (2mM each dNTP) 5μl 10×Pfu buffer 5μl Pfu DNA polymerase (10U / μl) 0.5μl ddH2O Bring the total volume of the reaction system to 50 μl
[0041] The prepared PCR reaction system was placed in the Bioer XP cycler gene amplification instrument, and the amplification was carried out according to the following p...
Embodiment 3
[0045] Through the method of whole gene synthesis, the secondary structure and codon bias of the gene are adjusted to achieve high expression in Escherichia coli.
[0046] Use Primer Premier (http: / / primer3.ut.ee / ) and OPTIMIZER (http: / / genomes.urv.es / OPTIMIZER / ) to design, and ensure that the Tm difference is controlled within 3°C, and the primer length is controlled within 60base , and the obtained primers were dissolved in double-distilled water, and added to the following reaction system, so that the final concentration of each primer was 30 nM, and the final concentration of the first and last primers was 0.6 μM.
[0047] 2mM dNTP mix (2mM each dNTP) 5μl 10×Pfu buffer 5μl Pfu DNA polymerase (10U / μl) 0.5μl wxya 2 o
Bring the total volume of the reaction system to 50 μl
[0048] The prepared PCR reaction system was placed in the Bioer XP cycler gene amplification instrument, and the amplification was carried out according to the foll...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com