A gene encoding protein factor rrf and its application in the production of n-acetylglucosamine
An acetamido, encoding gene technology, applied in the field of metabolic engineering, can solve problems such as undisclosed overexpression, and achieve the effects of increasing accumulation, good application prospects, and easy use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Example 1 Knockout of the L-lactate dehydrogenase encoding gene (ldh)
[0054] According to the upstream and downstream sequences of the Corynebacterium glutamicum ATCC 13032 L-lactate dehydrogenase coding gene (ldh) (nucleotide sequence shown in SEQ ID NO.1) published on NCBI, design knockout homology arms Amplification primers, left arm upper and lower primers are respectively LdhloxPUF (nucleotide sequence shown in SEQ ID NO.2) and LdhloxPUR (nucleotide sequence shown in SEQ ID NO.3); right arm upper and lower primers Respectively LdhloxPDF (nucleotide sequence as shown in SEQ ID NO.4) and LdhloxPDR (nucleotide sequence as shown in SEQ ID NO.5); Respectively with Corynebacterium glutamicum S9114 strain genomic DNA as template, by PCR Extend the left and right arms.
[0055] Primers KanloxpldhF (nucleotide sequence shown in SEQ ID NO.6) and KanloxpldhR (nucleotide sequence shown in SEQ ID NO.6) and KanloxpldhR (nucleotide sequence shown in Shown in SEQ ID NO.7), the...
Embodiment 2
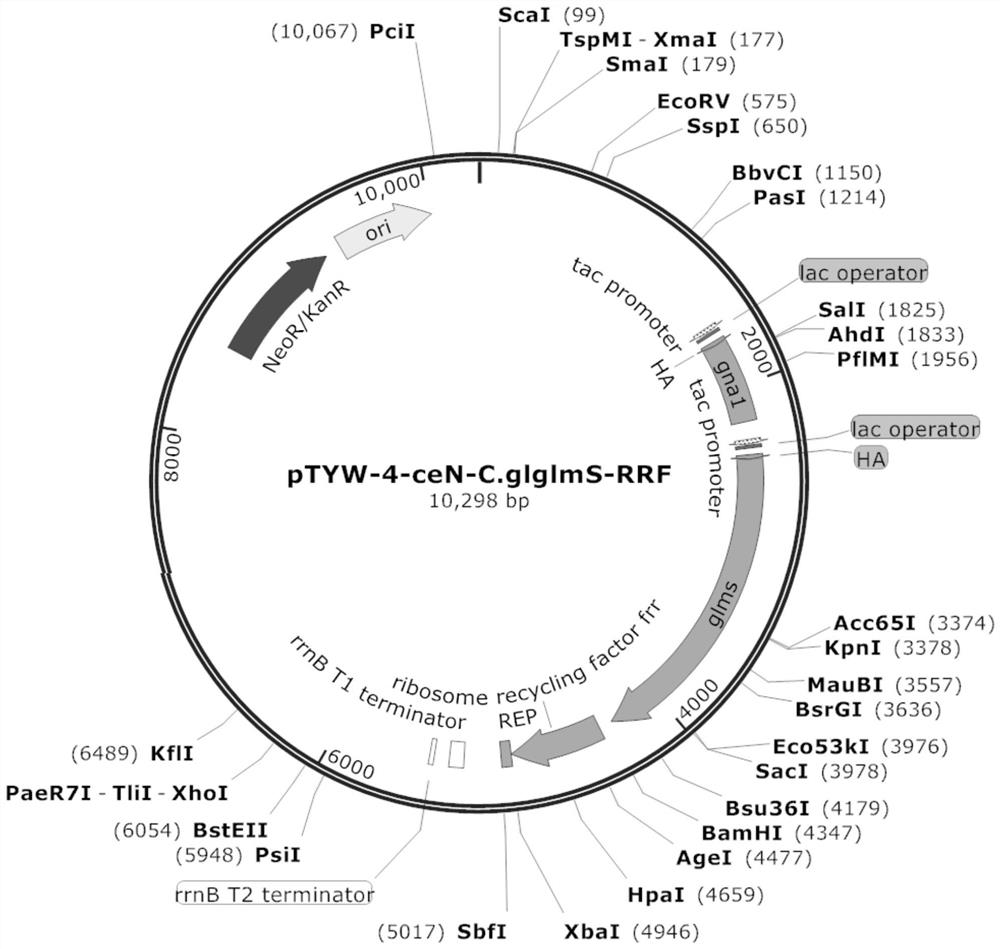
[0057] Example 2 Construction of recombinant plasmid pJYW-4-ceN-C.glglmS-RRF and construction of recombinant Corynebacterium glutamicum
[0058] (1) Design amplification primers to amplify RRF according to the S9114 genome
[0059] Upstream primer FragmentRRF.FOR:
[0060] 5' - ACCGTCGAATAATATAAACAGCGTGGCTTATCTAGGTTCG - 3';
[0061] Downstream primer FragmentRRF.REV:
[0062] 5' - CCTTTGCTAGTCTAGACCTCCTCATCAGTTCCTTTTTCCT - 3';
[0063] Simultaneously design vector pJYW-4-ceN-C.glglmS linearization primer
[0064] Upstream primer VectorRRF.FOR:
[0065] 5' - TGGAGGTCTAGACTAGCAAAGGAGAAGAAAAGCCG - 3';
[0066] Downstream primer VectorRRF.REV:
[0067] 5' - ACGCTGTTTATATTATTCGACGGTGACAGACTTTGC - 3';
[0068] Use primers FragmentRRF.FOR and FragmentRRF.REV, Corynebacterium glutamicum S9114 as a template, PCR conditions: 95°C for 10 min; 98°C for 1 min; 55°C for 1 min; 72°C for 1 min, 30 cycles of reaction; Finally, 72°C was extended for 10 min, and the PCR product was rec...
Embodiment 3
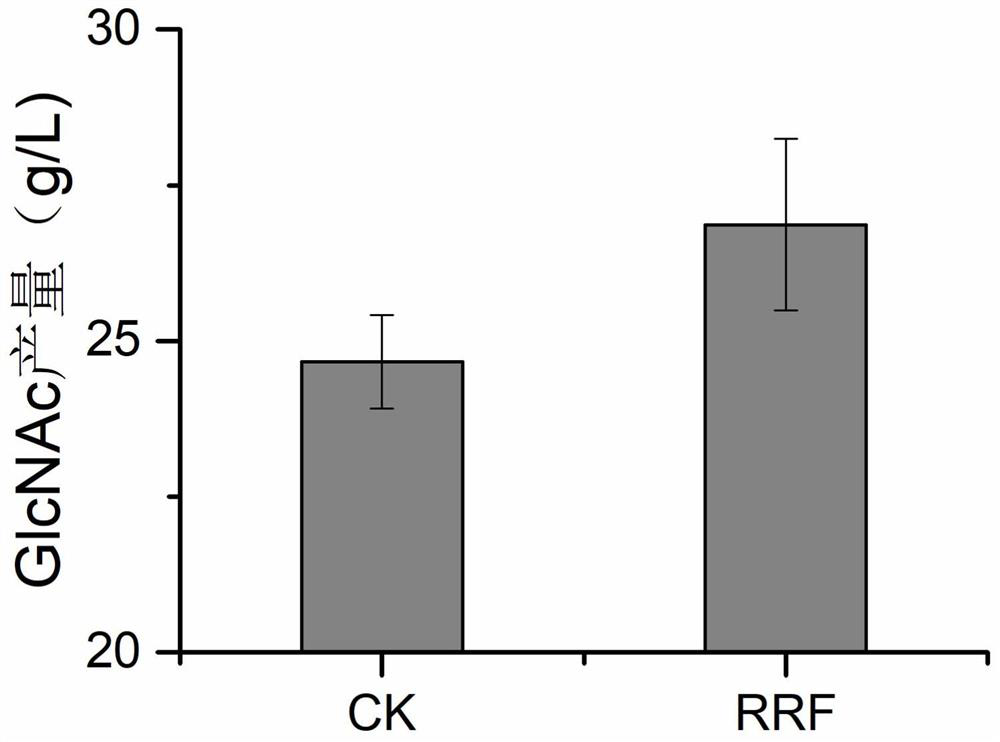
[0088] Effect of overexpressing frr gene on N-acetylglucosamine production in recombinant Corynebacterium glutamicum in embodiment 3
[0089] Inoculate the recombinant Corynebacterium glutamicum strain containing the plasmid pJYW-4-ceN-C.glglmS-RRF with the correct sequencing results on a streaked LBG plate (adding thiokanamycin 25 mg / L) in a glycerol tube, and inoculate at 30°C After culturing at 220rpm for 18 hours, select a single colony and re-stretch the LBG plate until a large number of colonies grow.
[0090] Inoculate a loop of single colonies into the seed medium, and culture at 220 rpm at 30°C for 16 to 18 hours until the cell growth logarithmic phase.
[0091] The seed culture solution was inoculated into the fermentation medium according to the inoculum amount of 10%, and cultivated at 30° C. and 220 rpm for 72 hours. GlcNAc production was measured.
[0092] Taking the recombinant bacterium containing plasmid pJYW-4-ceN-C.glglmS as a control, it was cultured and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com