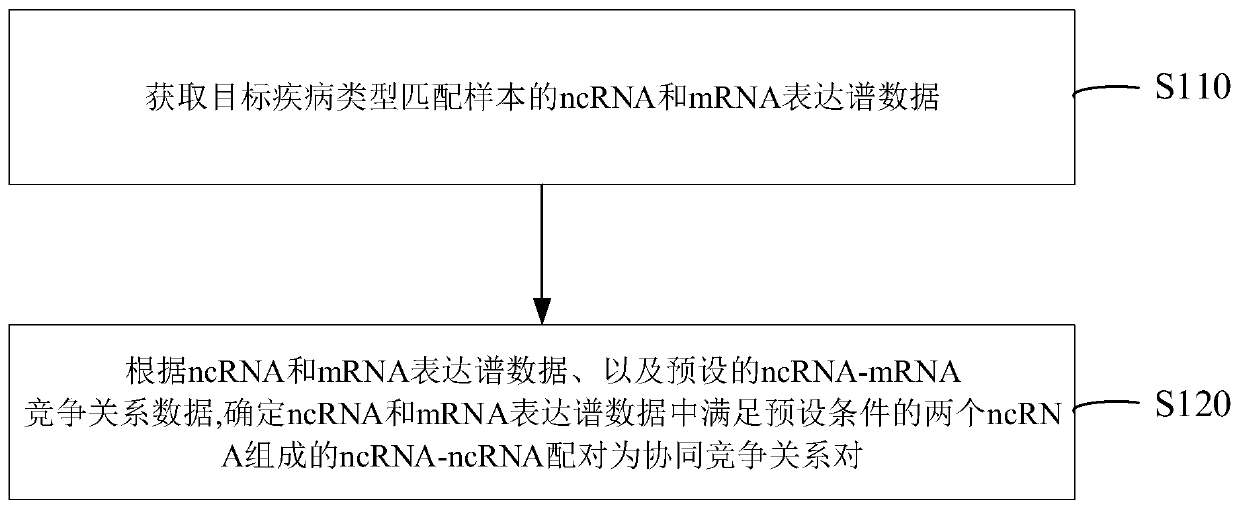
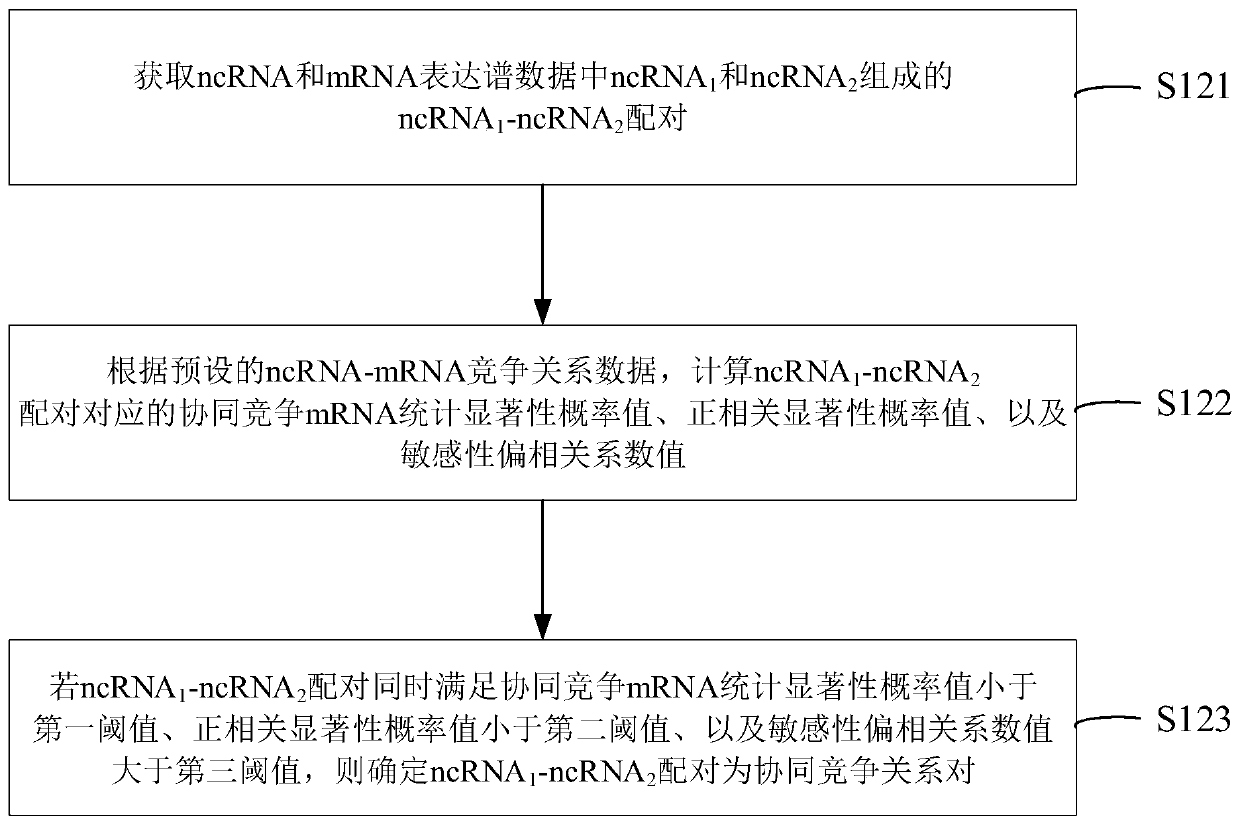
NcRNA collaborative competition network identification method and device
A technology for network identification and expression profiling, applied in instruments, for analyzing two-dimensional or three-dimensional molecular structures, hybridization, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0129] Taking the corresponding lncRNA-lncRNA cooperative competition in glioblastoma multiforme as an example, the method for identifying the lncRNA-lncRNA cooperative competition network in this embodiment is realized by the following steps:
[0130] Step 1: Data source acquisition
[0131] lncRNA and mRNA expression profile data of matched samples of glioblastoma multiforme (GBM) were collected from the internationally renowned cancer gene expression profile database TCGA (the cancer genome atlas, https: / / cancergenome.nih.gov / ) . Through preprocessing (removing duplicates and lncRNAs and mRNAs without gene names), the expression profile data of 9704 lncRNAs and 18282 mRNAs of 451 breast cancer matched samples, as well as the clinical information of the samples were finally obtained. In this embodiment, ncRNA is lncRNA,
[0132] The prior lncRNA-mRNA competition network data was obtained by fusing multiple different databases. Specifically, by integrating miRSponge, L...
Embodiment 2
[0162] Taking the corresponding lncRNA-lncRNA cooperative competition in lung squamous cell carcinoma as an example, the method for identifying the lncRNA-lncRNA cooperative competition network in this embodiment is realized by the following steps:
[0163] In step 1 of this embodiment, the lncRNA of the matched sample of lung squamous cell carcinoma (Lung Squamous Cell Carcinoma, LSCC) was collected from the internationally renowned cancer gene expression profile database TCGA (the cancergenomeatlas, https: / / cancergenome.nih.gov / ) and mRNA expression profile data. After preprocessing (removing duplicates and lncRNAs and mRNAs without gene names), the expression profile data of 9704 lncRNAs and 18282 mRNAs of 113 breast cancer matched samples, as well as the clinical information of the samples were finally obtained. In this embodiment, ncRNA is lncRNA,
[0164] The prior lncRNA-mRNA competition network data is the same as in Example 1, and finally 10099 lncRNA-mRNA compet...
Embodiment 3
[0167] Taking the corresponding lncRNA-lncRNA cooperative competition in ovarian cancer as an example, the method for identifying lncRNA-lncRNA cooperative competition network in this embodiment is realized by the following steps:
[0168] In step 1 of this example, lncRNA and mRNA expressions of ovarian cancer (Ovarian Cancer, OvCa) matched samples were collected from the internationally renowned cancer gene expression profile database TCGA (the cancergenome atlas, https: / / cancergenome.nih.gov / ) spectral data. Through preprocessing (removing duplicates and lncRNAs and mRNAs without gene names), the expression profile data of 9704 lncRNAs and 18282 mRNAs of 585 ovarian cancer matched samples, as well as the clinical information of the samples were finally obtained. In this embodiment, ncRNA is lncRNA,
[0169] The prior lncRNA-mRNA competition network data is the same as in Example 1, and 10099 lncRNA-mRNA competition relationship pairs associated with ovarian cancer expr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com