Method and device for screening candidate drugs for COVID-19
A technology of medicine and medicinal chemistry, applied in bioinformatics, instrumentation, genomics, etc., can solve problems such as new drugs or vaccines with little practical significance, and achieve the effect of accelerating drug development speed, high precision, and reducing drug development costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
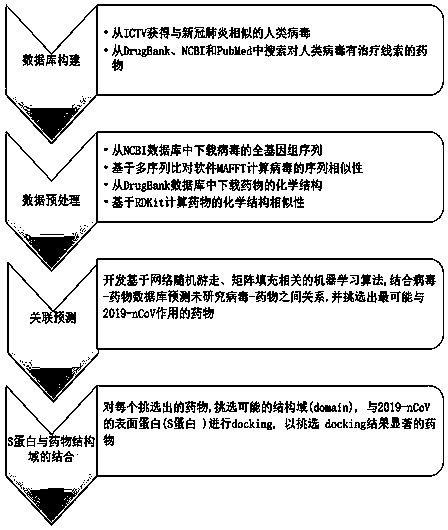
[0076] 1. Sample information
[0077] The whole genome sequences of 12 RNA viruses from NCBI database and the chemical structures of 78 drugs from DrugBank database and PubMed database were selected as samples.
[0078] 2. Experimental steps
[0079] 1. Preprocessing:
[0080] 1.1 Mining viruses similar to 2019-nCoV from ICTV virus data, the initial statistics are 11 kinds, including influenza viruses including A-H1N1, A-H5N1 and A-H7N9, chronic hepatitis C virus (HCV), HIV-1 and HIV-2, Hendra virus, human cytomegalovirus, Middle East respiratory syndrome coronavirus (MERS virus), severe acute respiratory syndrome (SARS) virus, and respiratory syncytial virus.
[0081] 1.2 Download the associated drugs of 11 viruses from the DrugBank database.
[0082] 1.3 From the PubMed database to search for drugs associated with 11 viruses.
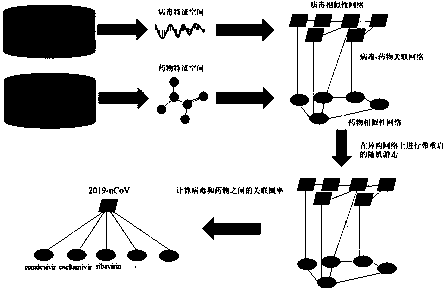
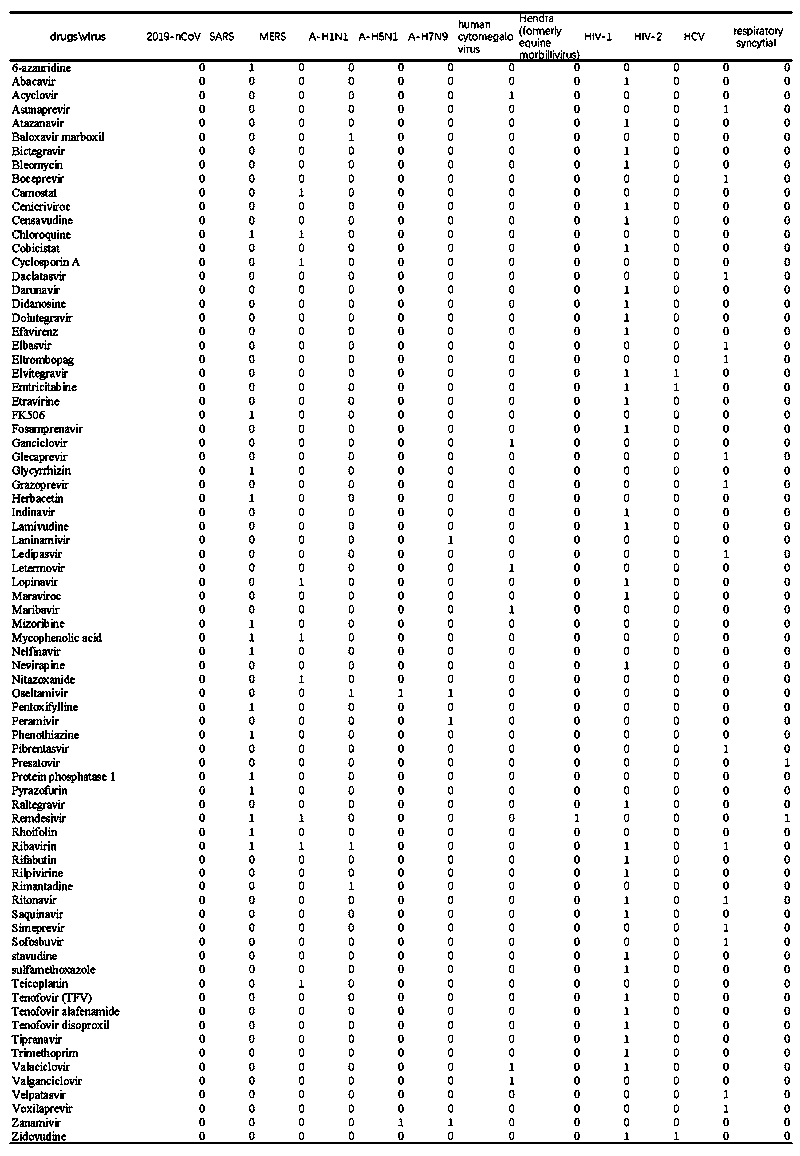
[0083] 1.4 Construct virus-drug association database, and build image 3 matrix shown.
[0084] Similarity calculation:
[0085] 2.1 Download ...
Embodiment 2
[0115] The other steps of Example 2 are the same as those of Example 1 unless otherwise specified. The sample information in this embodiment is to select the whole genome sequences of 11 RNA viruses from the NCBI database and the chemical structures of 63 drugs from the DrugBank database and the PubMed database as samples. Among them, the viruses are SARS virus and its similar viruses A-H1N1, A-H5N1, A-H7N9, HCV, HIV-1, HIV-2, Hendra virus, human cytomegalovirus, MERS virus and respiratory syncytial virus.
[0116] In this embodiment, after determining the performance of the model, SARS is regarded as a new virus, and the drug information associated with it is completely covered up, and the random walk algorithm with restart is used to predict the drug associated with SARS. Table 2-4 shows the 63 drugs associated with SARS and their rankings. All of these small molecule drugs are FDA-approved.
[0117] Table 2-4 Drugs and their rankings associated with SARS
[0118]
[0...
Embodiment 3
[0122] The other steps of Example 3 are the same as those of Example 1 unless otherwise specified.
[0123] The sample information in this embodiment is to select the whole genome sequences of 11 viruses from the NCBI database and the chemical structures of 69 drugs from the DrugBank database and the PubMed database as samples. The viruses are MERS virus and its similar viruses A-H1N1, A-H5N1, A-H7N9, HCV, HIV-1, HIV-2, Hendra virus, human cytomegalovirus, SARS virus and respiratory syncytial virus.
[0124] In this example, after determining the performance of the model, MERS is regarded as a new virus, and the drug information associated with it is completely covered up, and the random walk algorithm with restart is used to predict the drugs associated with MERS. Table 3-4 shows the 69 drugs associated with SARS and their rankings. All of these small molecule drugs are FDA-approved.
[0125] Table 3-4 Drugs and their rankings associated with MERS
[0126]
[0127] In t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com