Test kit and detection method for quantitative detection of high-throughput sequencing library
A sequencing library and quantitative detection technology, applied in the field of high-throughput sequencing, can solve problems such as uneven sequencing data quality, and achieve the effect of improving sequencing quality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
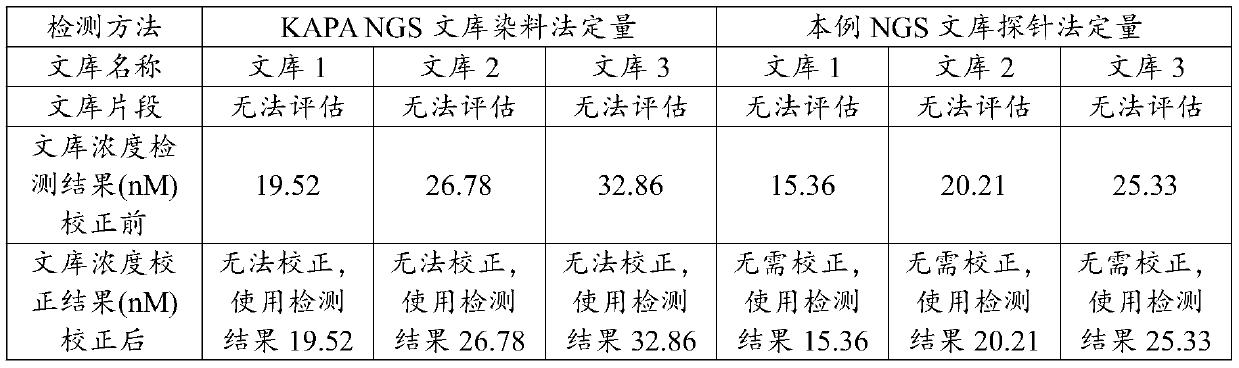
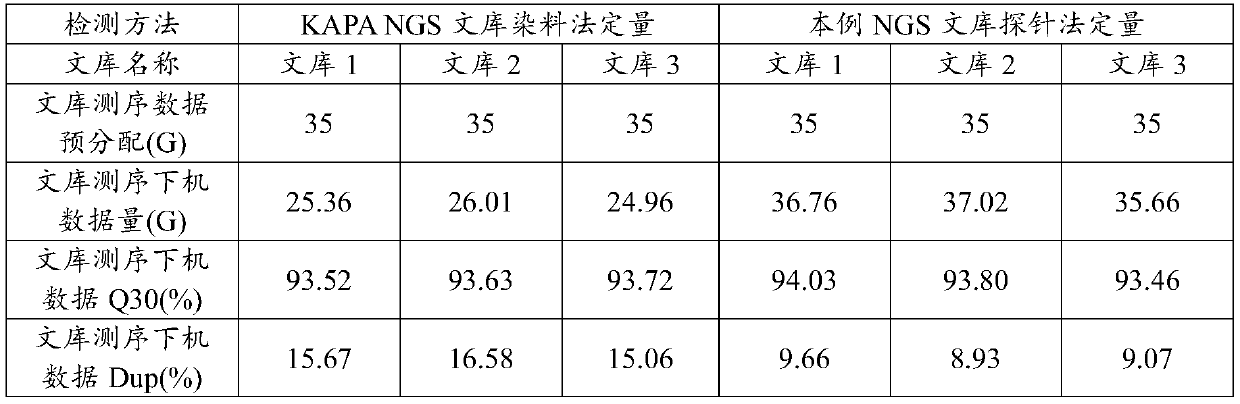
[0029] In this example, human leukocyte genomic DNA (gDNA) was used to construct a high-throughput sequencing library based on the Illumina sequencing platform. Specifically, KAPAHyper Prep Kit (KK850407962363001) was used to construct a high-throughput sequencing library of three human leukocyte gDNA. The specific steps of high-throughput sequencing library construction refer to the instruction manual of the KAPHyperPrep Kit kit. The three high-throughput sequencing libraries constructed by human leukocyte gDNA are labeled as library 1, library 2, and library 3.
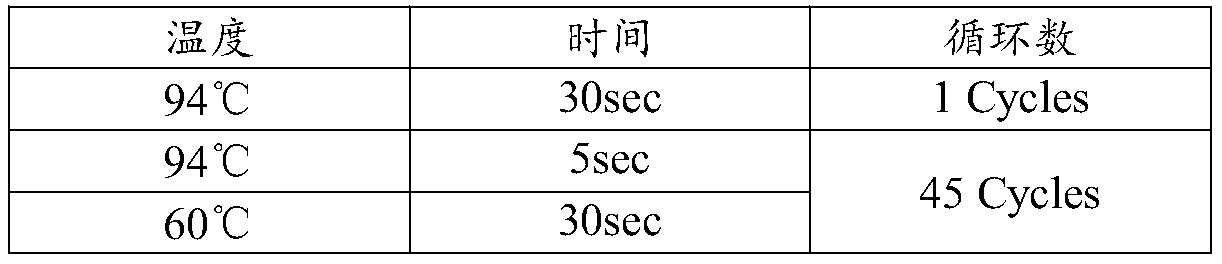
[0030] Among them, the gDNA sample fragmentation was performed using an ultrasonic disintegrator Covaris M220. The specific parameters are: 500μL of gDNA, which is 1000ng, temperature 20°C, Peak Power 50.0, Duty Factor 30.0, Cycle / Burst 200, and time 300sec.
[0031] For the adapter sequence of the Illumina sequencing platform, detection primers and detection probes for the adapter sequence are designed in this example; ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com