Blood mRNA detection kit and detection method
A technology for detection kits and blood, applied in the directions of DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve allele loss, does not solve complex mixed plaques, and cannot break through the detection sensitivity of complex mixed plaques. Bottlenecks and other problems to achieve the effect of perfect separation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
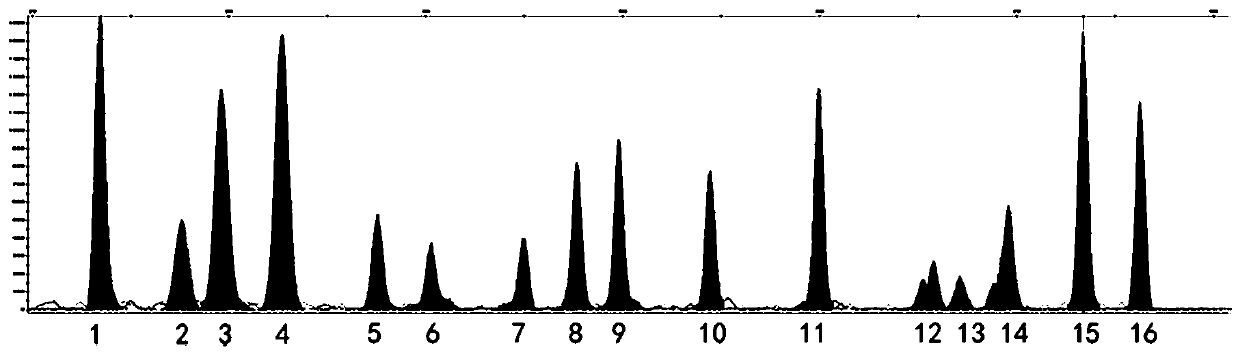
[0033] Example 1. Screening of mixed plaques.
[0034] 1. Prepare the template.
[0035] Whole blood RNA was extracted using the RNAiso Plus kit (Takara). After quantification, 100 ng of RNA was used as a template for reverse transcription using the GoScript Reverse Transcription Kit (Promega). Obtain template cDNA.
[0036] 2. Compound amplification and purification.
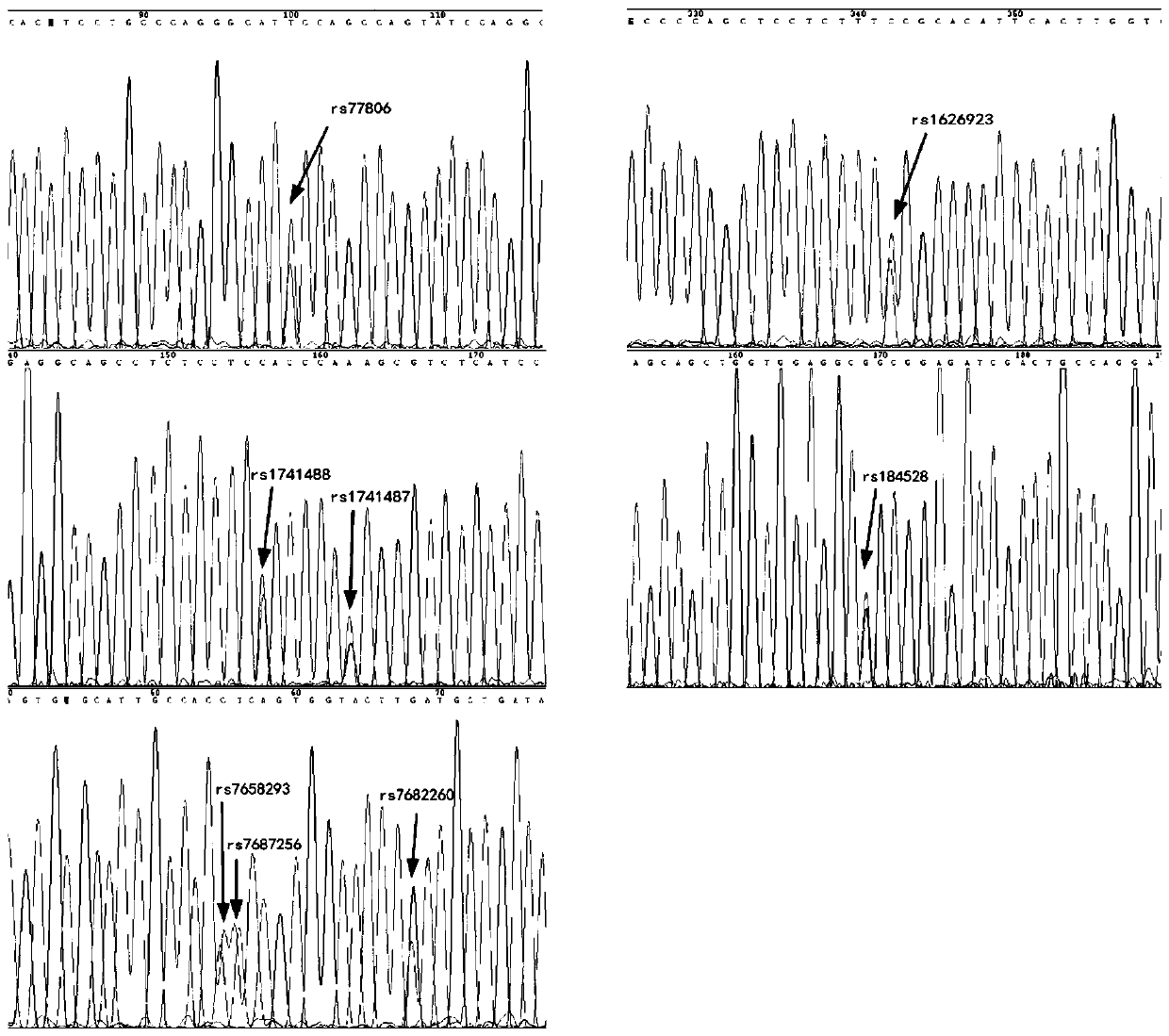
[0037] 1) Add template cDNA representing 5ng RNA to 10 μL reaction system: 5 μL 2×Mastermix; 1 μL 10×Primer Mix (see Table 2 for primer concentration), make up the reaction system to 10 μL with water. The PCR thermocycling conditions are as follows: 95°C for 10 min; 30 cycles of 95°C for 30s, 60°C for 30s, and 72°C for 20s; 72°C for 5min.
[0038]2) Purification system for complex amplification product: 3.8 μL of amplification product, 1.3U SAP (shrimp alkaline phosphatase), 6U Exo Ι (exonuclease Ι), make up the reaction system to 10 μL with water. Purification conditions: 37°C, 1h; 95°C, 15min.
[0039] ...
Embodiment 2
[0048] Example 2. Screening of old plaques.
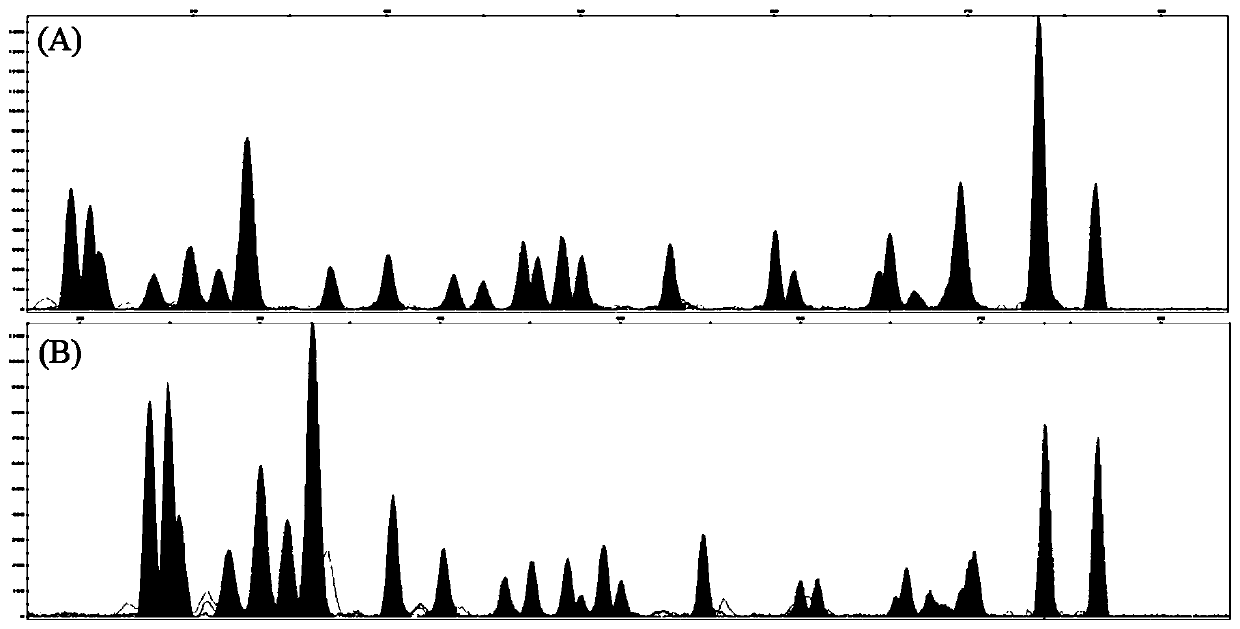
[0049] Use this kit to use the blood spot sample stored at room temperature for 6 months as a template for detection, the results are shown in Figure 4 , and all typing results were obtained.
Embodiment 3
[0050] Example 3. Screening of extremely unbalanced mixed plaques.
[0051] Use this kit to amplify and SNaPshot type the template containing 0.1ng RNA, the results are shown in Figure 5 , and also obtained all typing results.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com