Method for carrying out blocking, substitution, amplification, enrichment and detection on target mutation based on blocker introducing extra base mismatch
A technology for detecting targets and bases, which can be used in biochemical equipment and methods, determination/inspection of microorganisms, etc., and can solve problems such as inability to meet requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
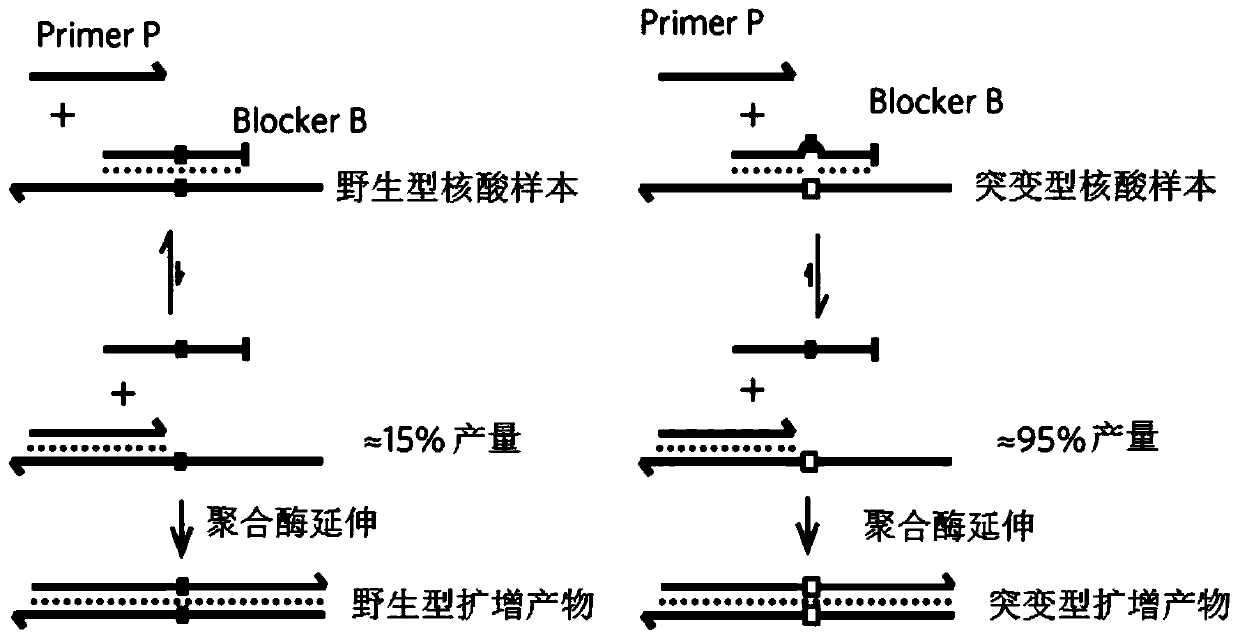
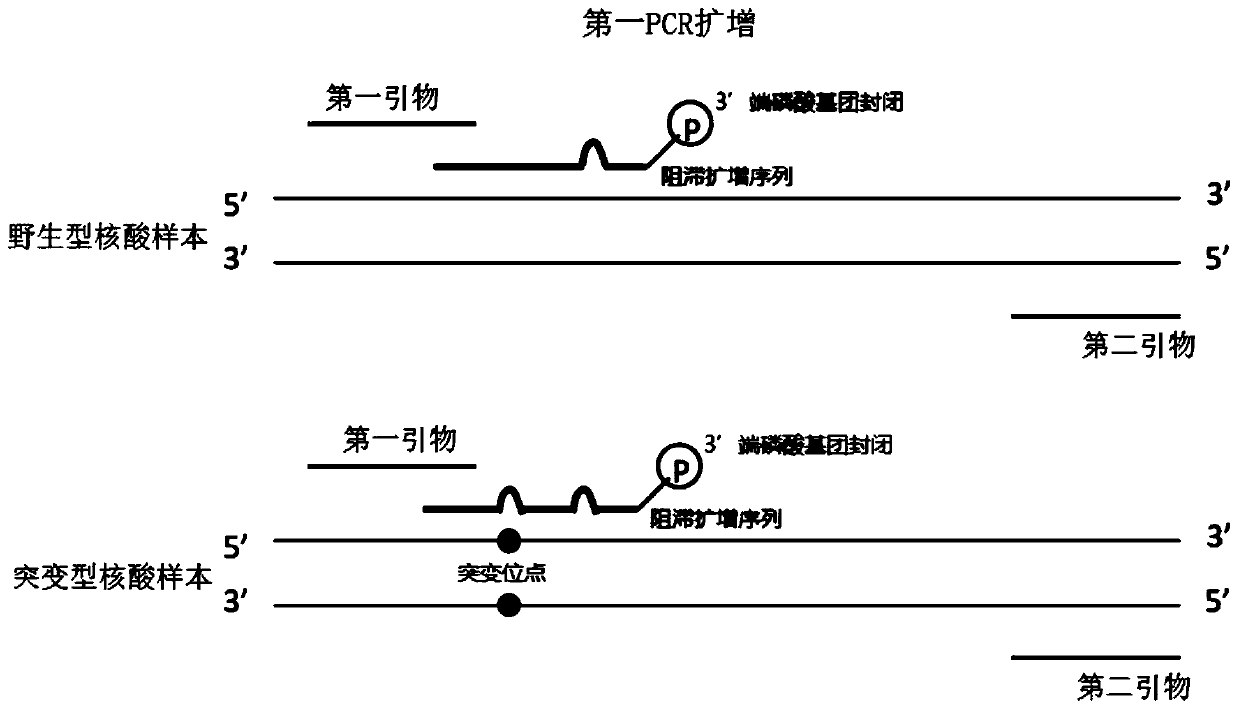
[0053] Example 1 provides a method for enriching and detecting the target region, and takes the detection of EGFR G719S mutation as an example to illustrate. The sample DNA used is: wild-type cfDNA and 1% mutant cfDNA (referring to sample DNA The mutated sequence containing the target mutation site accounts for 1% of the total sequence).
[0054] Including the following steps:
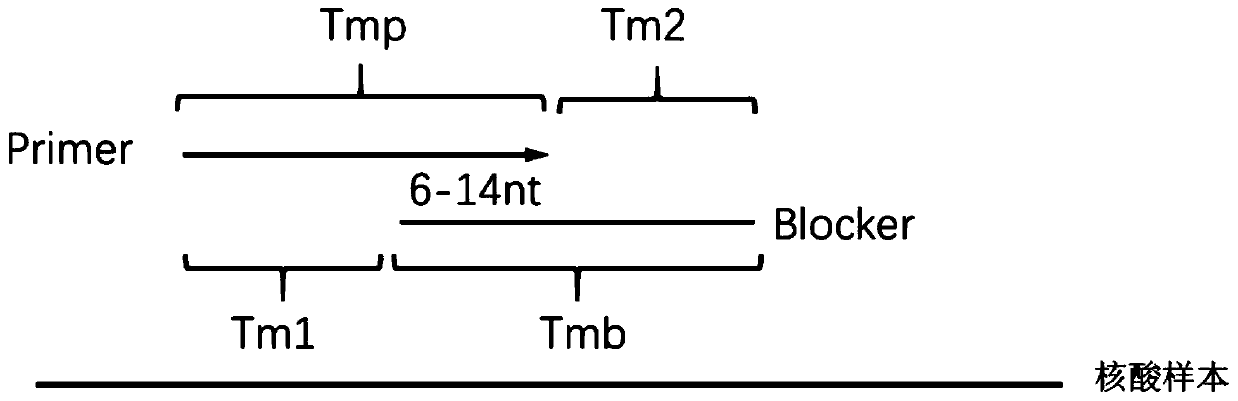
[0055] 1. Using the first primer, the second primer and the blocking amplification sequence to perform the first PCR amplification.
[0056] First prepare the following PCR system:
[0057] Table 1 The first PCR system
[0058]
[0059]
[0060]Among them, 2X Thermo Enzyme Mix comes from Platinum SuperFi PCR Master Mix, the manufacturer is ThermoFishier Scientific, and the article number is 00772786.
[0061] Perform the first PCR amplification according to the following PCR program:
[0062]
[0063] The primer sequences and retardation amplification sequences used were artificially synth...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com