Method for detecting target mutation by carrying out retardation substitution amplification enrichment based on locked nucleic acid modified blocker
A detection target and enrichment technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve problems such as unsatisfactory requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
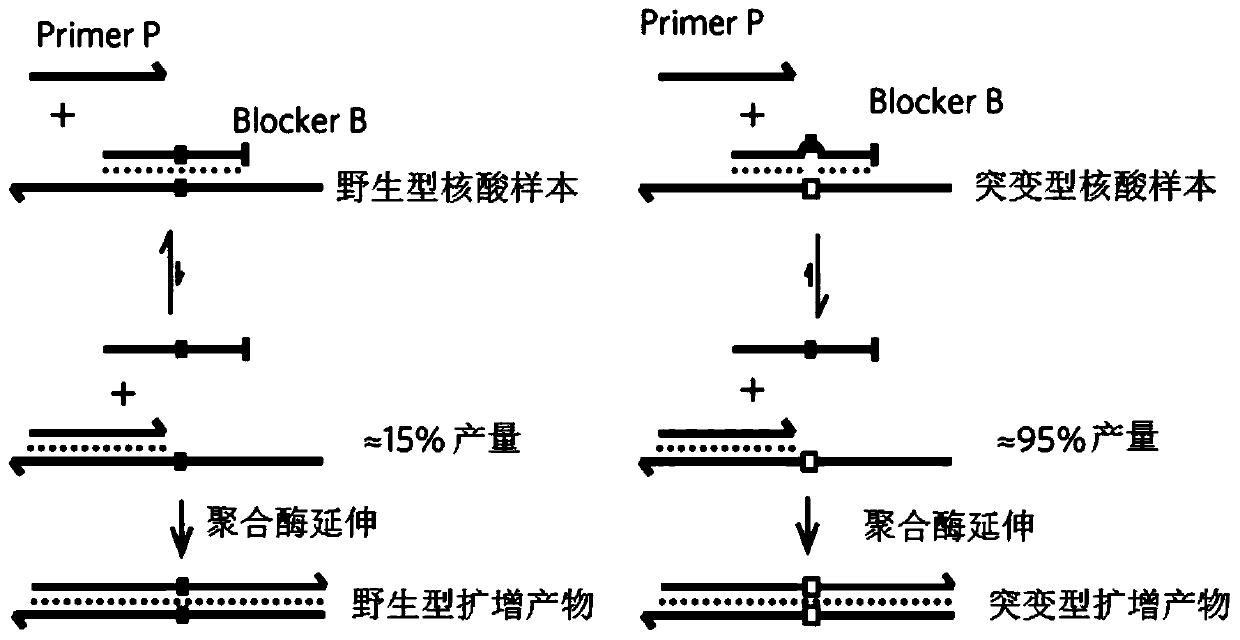
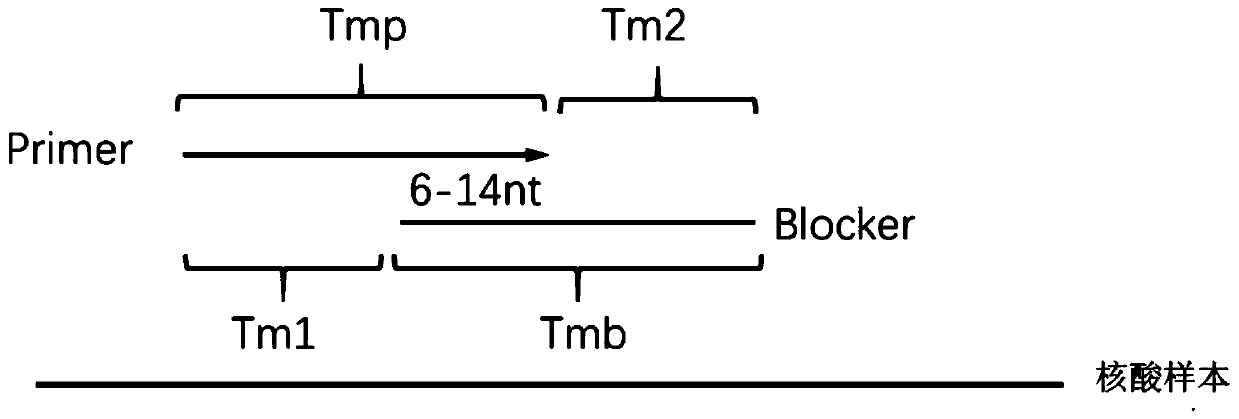
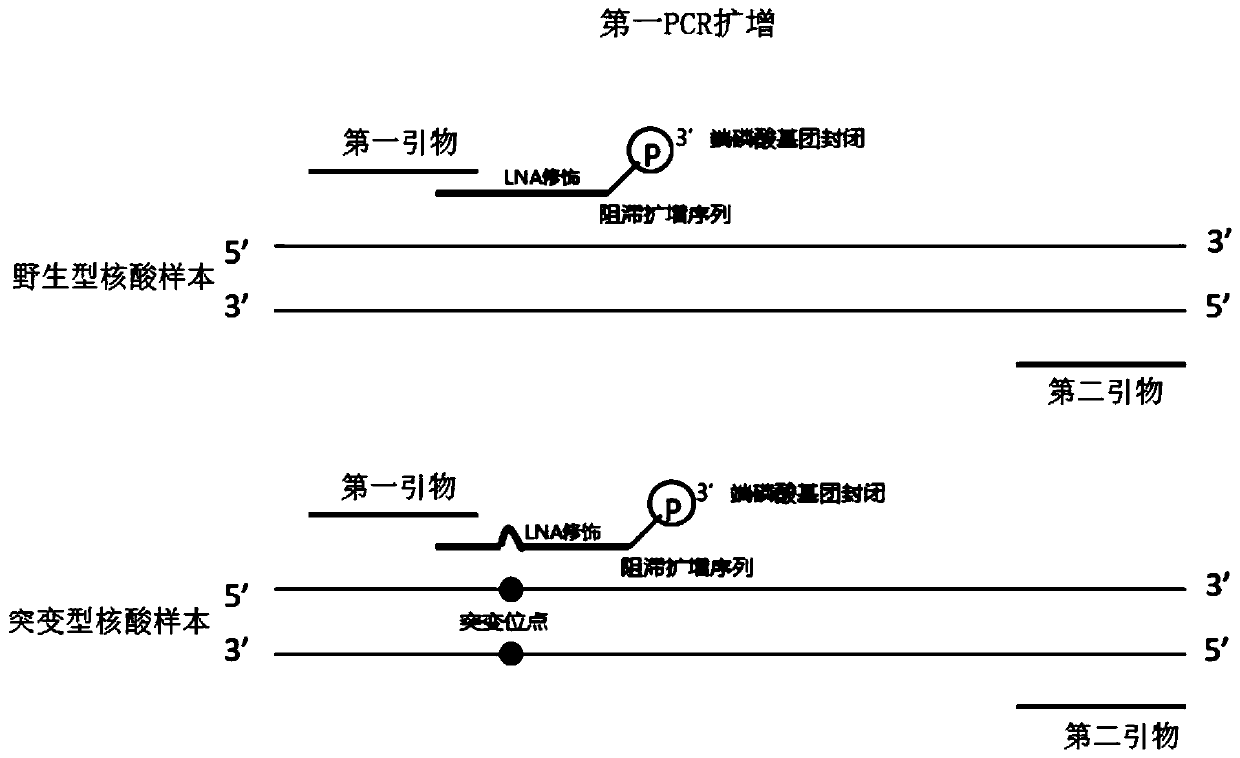
[0053] Example 1 provides a method for enriching and detecting the target region, and takes the detection of EGFR L858R mutation as an example to illustrate. The sample DNA used is: wild-type cfDNA and 0.1% mutant cfDNA (referring to sample DNA The mutated sequence containing the target mutation site accounts for 0.1% of the total sequence), including the following steps:
[0054] 1. Use the first primer, the second primer and the retardation amplification sequence to carry out the first PCR amplification
[0055] Mix and amplify according to the following reaction system and reaction procedures.
[0056] Table 1 The first PCR system:
[0057] Reagent volume Sample DNA amount (20ng / μL) 1μL First primer (10 μM) 1μL Second primer (10 μM) 1μL EGFR L858R blocking amplification sequence (100μM) 2.5μL wxya 2 O (μL)
19.5μL 2X Thermo Enzyme Mix (μL) 25 μL Total volume (μL) 50μL
[0058] Among them, 2X Thermo Enzyme...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com