Methylation molecular markers for diagnosing lung adenocarcinoma and application of methylation molecular markers for diagnosing lung adenocarcinoma
A molecular marker and methylation technology, applied in the field of biomedical detection, can solve problems such as the survival rate of less than 20%, unsatisfactory survival of lung adenocarcinoma patients, and no report on the EID3 gene, achieving high detection accuracy, Accurately detect the effect of the method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 TCGA methylation database research
[0027] 1. Data download:
[0028] The TCGA lung adenocarcinoma methylation database was downloaded from the official website of the TCGA database at https: / / tcga-data.nci.nih.gov / tcga / , and the search keywords were lung adenocarcinoma and DNA methylation. The data download date is 2019-03-27. All data were normalized and the total sample size of the DNA methylation cohort was 616 (53 normal, 563 lung adenocarcinoma).
[0029] 2. Data processing:
[0030] (1) All data were normalized before further analysis.
[0031] (2) Chi-square test was used to analyze the difference of gene methylation in lung adenocarcinoma and adjacent lung tissue.
[0032] (3) Differentially methylated genes were screened using p=0.05 and |logFc|=0.5 as criteria.
[0033] (4) The TCGA data download software is Perl, and the software used for data processing and analysis is R with version number 3.3.1.
[0034] 3. Data analysis results:
[0035...
Embodiment 2
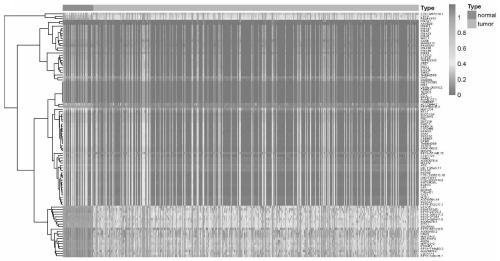
[0035] In the present invention, 616 sample information were included in the TCGA gene methylation cohort, including 563 lung adenocarcinoma cases and 53 control cases. We used p-value 0.5 for screening, and found that among the 20204 genes with methylation data, a total of 2084 genes were differentially methylated between lung adenocarcinoma tissue and paracancerous tissue, of which There were 369 hypermethylated tumor tissues and 1715 relatively hypomethylated tumor tissues compared with paracancerous tissues. We show a heat map of the top 100 differentially methylated genes ( figure 1 ). Example 2 Quantitative study on tissue EID3 methylation level
[0036] 1. Research object:
[0037] The 30 cases of lung adenocarcinoma and adjacent normal lung tissue samples we used were from patients who were diagnosed with lung adenocarcinoma by the Department of Thoracic Surgery of East China Hospital and underwent surgical resection. Immediately after resection, the obtained tissu...
Embodiment 3
[0097] Embodiment 3: clinical validation
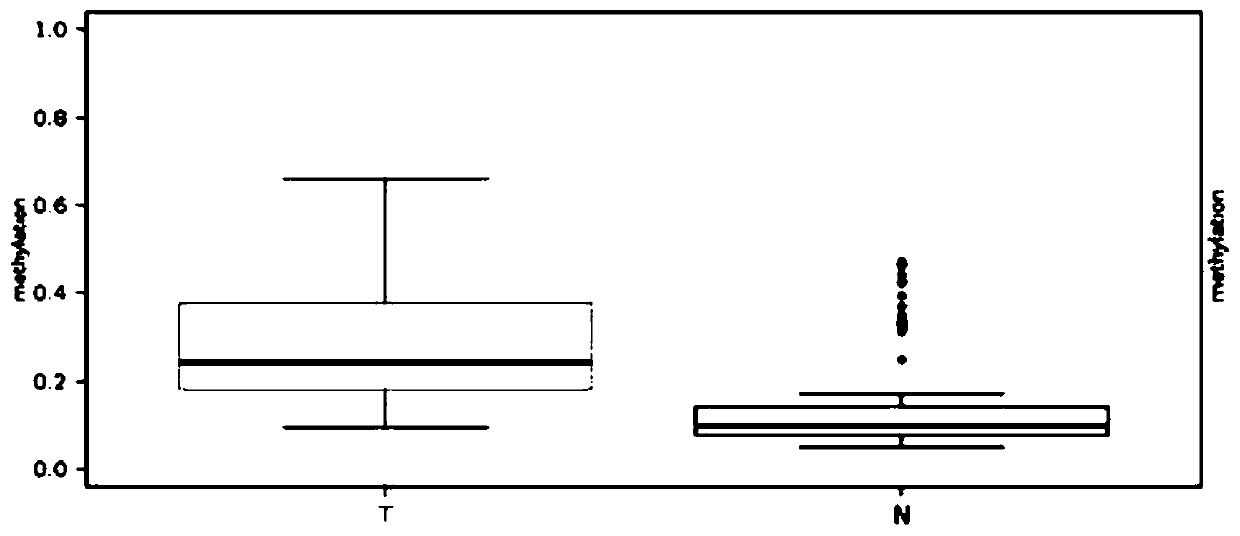
[0098] According to the above method, we detected the methylation level of EID3 gene in 30 samples (including lung adenocarcinoma tissue and adjacent normal tissue) in East China Hospital, and further analyzed the methylation level of each methylation site included in the tumor and adjacent normal tissue. difference in methylation. The inventors found that, in East China Hospital samples, EID3 showed significant hypermethylation in the lung adenocarcinoma group relative to the paracancerous tissues ( image 3 ). And among the detected methylation sites, except for 194, 204, and 208 sites, the methylation levels of other sites are relatively low. The methylation of all sites in the T group was significantly higher than that in the N group ( Figure 4 ). Through cluster analysis, the present inventors found that in lung adenocarcinoma and adjacent tissues, there is a significant difference in the level of RHOH methylation, and it ca...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com