Nucleic acid aptamer for specifically recognizing bovine pregnancy-associated glycoproteins and application of nucleic acid aptamer
A nucleic acid aptamer and glycoprotein technology, which is applied in the fields of biochemical equipment and methods, material testing products, instruments, etc., can solve the problems such as no research reports on livestock pregnancy-related glycoprotein nucleic acid aptamers, etc., and achieve good affinity. and specificity, short production cycle, easy chemical modification effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1: Screening of bPAG nucleic acid aptamers
[0031] 1. Synthetic random single-stranded DNA (ssDNA) library and primers:
[0032] Random single-stranded DNA (ssDNA) library:
[0033] 5'-CTACGGTGCCTTGAAGTGAC-N36-CATAGCAGGTCACTTCCAGG-3', wherein, N36 represents 36 random nucleotides, and the library was synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.;
[0034] Upstream primer: 5'-FAM-CTACGGTGCCTTGAAGTGAC-3',
[0035] Downstream primer: 5'- 20A-spacer18-CCTGGAAGTGACCTGCTATG-3',
[0036] Among them, in the downstream primers, 20A represents a polyA tail composed of 20 adenosine (A), and Spacer 18 represents an 18-atom hexaethylene glycol interarm. The above primers were synthesized by Nanjing GenScript Biotechnology Co., Ltd.
[0037] The random single-stranded DNA library, 5' end primer, and 3' end primer were respectively washed with PBS buffer (PBS, NaCl: 8 g / L, KCl: 0.2 g / L, NaCl: 0.2 g / L, Na 2 HPO 4 : 1.15 g / L, KH 2 pH 4 : 0.2 g / L; PH 7 .4) dis...
Embodiment 2
[0049] Embodiment 2: SPR detects the dissociation constant of nucleic acid aptamer and bPAG9 protein
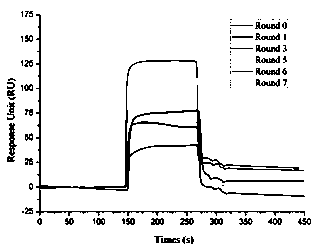
[0050] Take several pieces of ssDNA synthesized in Example 1, that is, nucleic acid aptamers, and each nucleic acid aptamer is prepared into a series of gradient concentration aptamer solutions with PBS; couple bPAG9 protein on the chip according to step 4, and then mix different concentrations of The aptamer solution is injected into the chip coupled with bPAG9 sequentially, and the affinity of each nucleic acid aptamer and bPAG9 protein is detected, and it is found that the nucleic acid aptamer of the sequence shown in SEQ ID No.1 and SEQ ID No.2 has the same affinity with the bPAG9 protein High affinity, the dissociation constants obtained by SPR detection are all in nM level (see image 3 ); wherein, sequence 1 is 1.04 nM for the dissociation constant of the nucleic acid aptamer of the sequence shown in SEQ ID No.1 to the bPAG9 protein, and sequence 2 is the nucleic acid ...
Embodiment 3
[0051] Embodiment 3: SPR detects the binding specificity of nucleic acid aptamer and bPAG protein
[0052] Dilute the four similar proteins of bPAG4, bPAG6, bPAG9 and bPAG16, as well as BSA and OVA proteins with sodium acetate solution (pH5.0) to a concentration of 50 μg / mL, and couple the proteins on different channels of the chip according to step 4. The nucleic acid aptamer sequences shown in SEQ ID No. 1 and SEQ ID No. 2 were all prepared in PBS to a concentration of 500 nM, and the binding of each nucleic acid aptamer to each protein was determined according to step 4. Test results such as Figure 5 , the nucleic acid aptamers shown in SEQ IDNo.1 and SEQ IDNo.2 can all be combined with b-PAG 4, b-PAG 6, b-PAG 9, b-PAG 16 in the bPAG protein family, but not with BSA Binds to OVA protein.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com