Multi-scale CNN-BiLSTM non-coding RNA interaction relationship prediction method with introducing attention
A prediction method and attention technology, applied in the fields of bioinformatics and deep learning, can solve problems such as large feature matrix, increased memory and calculation burden, and loss of effective information
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
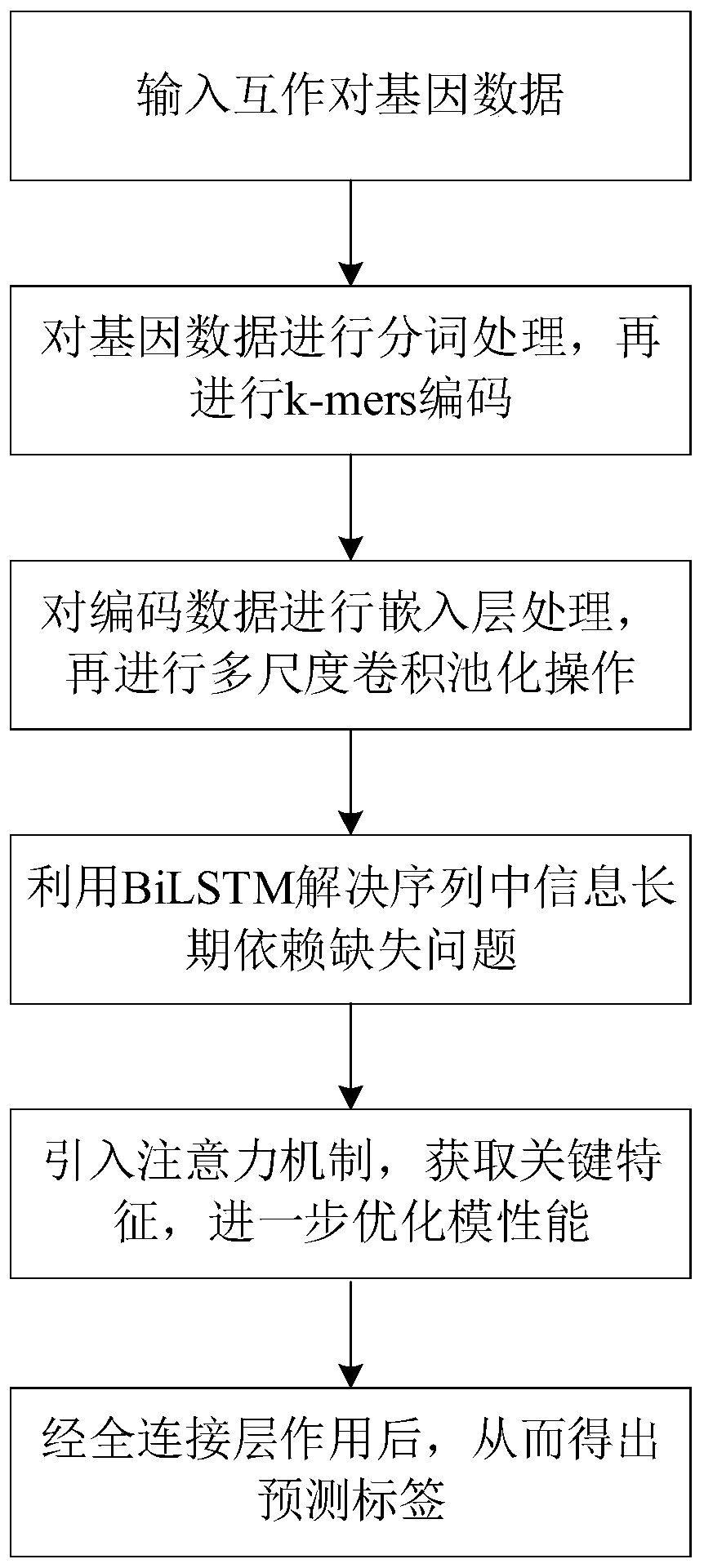
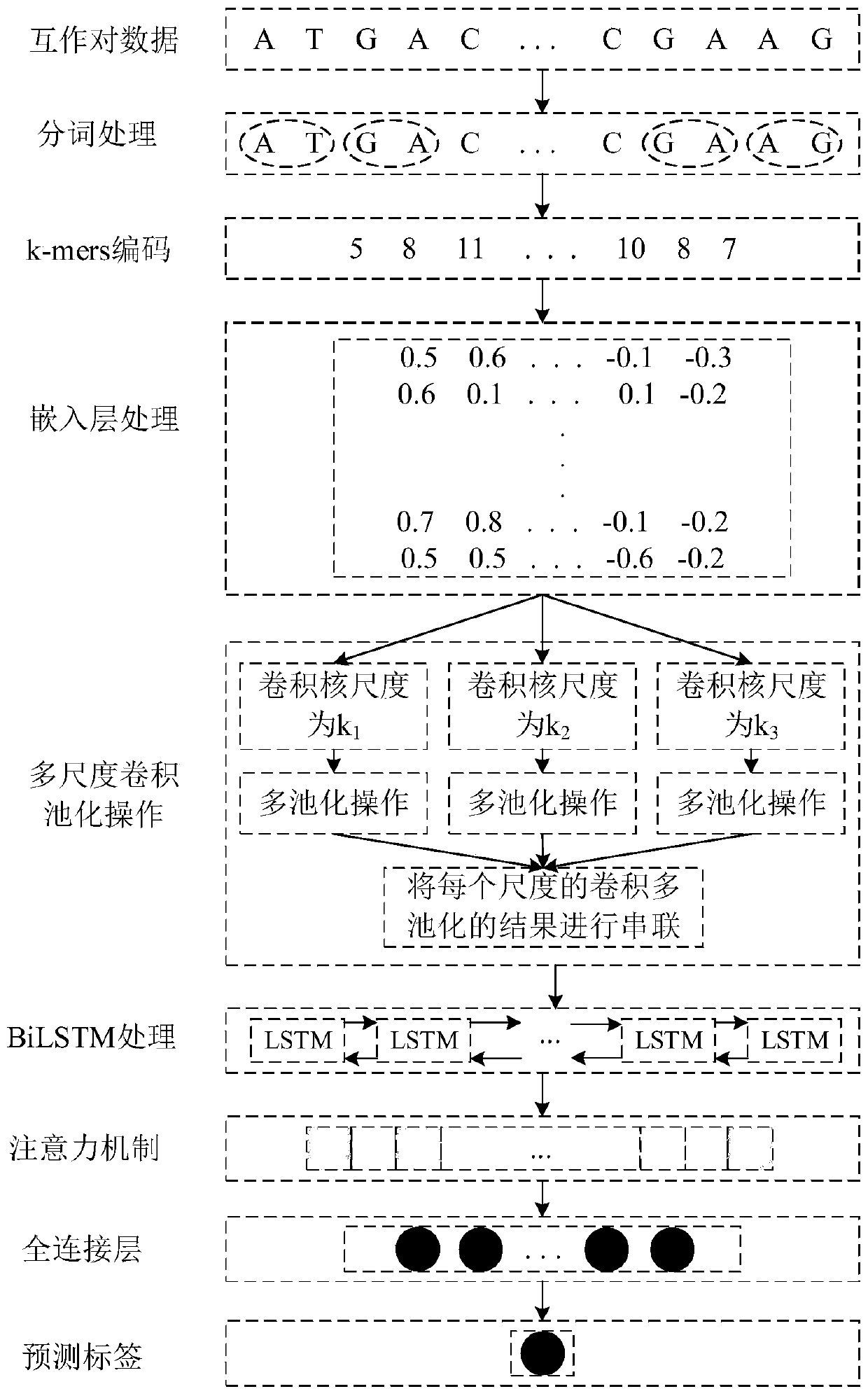
[0067] The specific implementation manners of the present invention will be further described below in conjunction with the accompanying drawings and technical solutions.
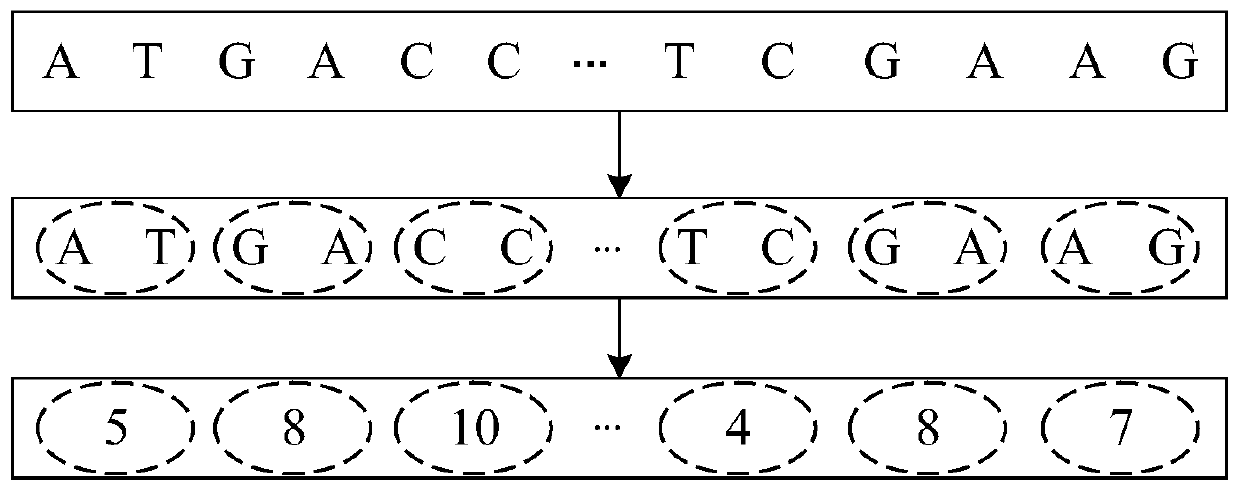
[0068] Such as figure 2 As shown, the overall design idea of the present invention is: because the traditional coding method does not consider the dependent relationship between adjacent nucleotides, in fact, there is a certain dependent relationship between adjacent nucleotides, therefore, the gene The sequence is encoded by k-mers, which preserves the dependent relationship between adjacent nucleotides and avoids the loss of potential effective information; then uses the embedding layer to map the encoded sequence into a matrix vector for convolution operations, and uses multi-scale convolution and Multi-pool operation replaces single-scale convolution and single-pool operation, so that features of different topic lengths can be extracted, and feature diversity can be enriched to achieve the purpose of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com