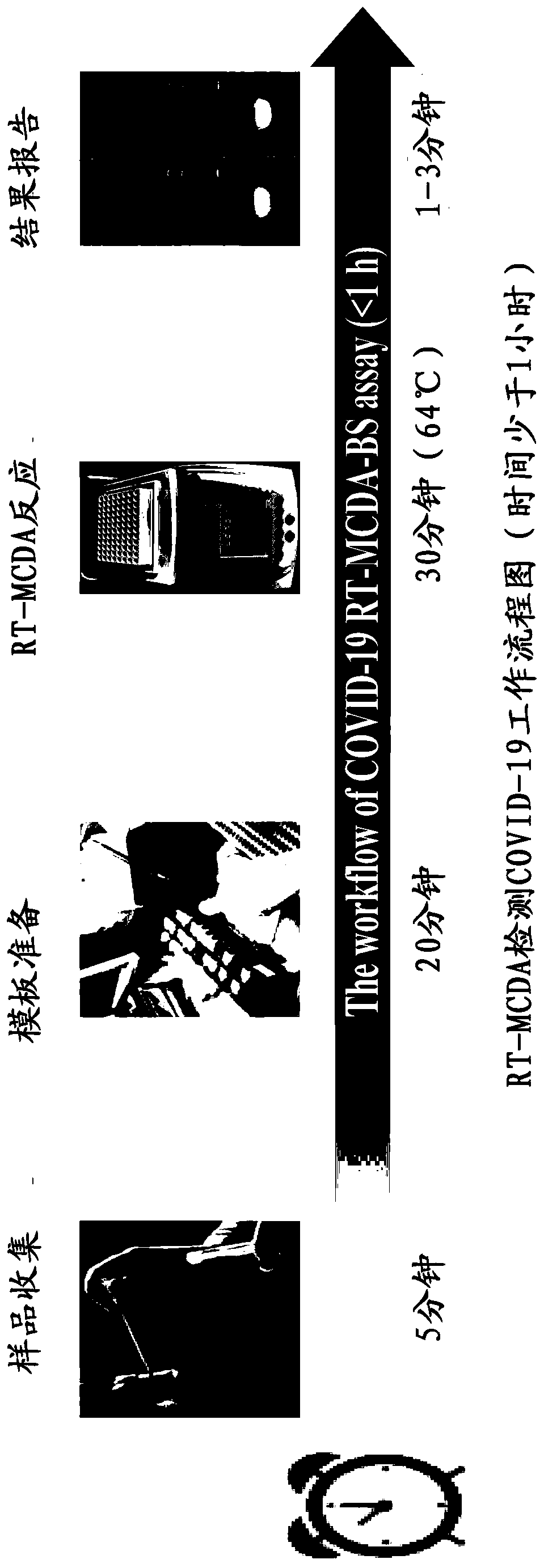
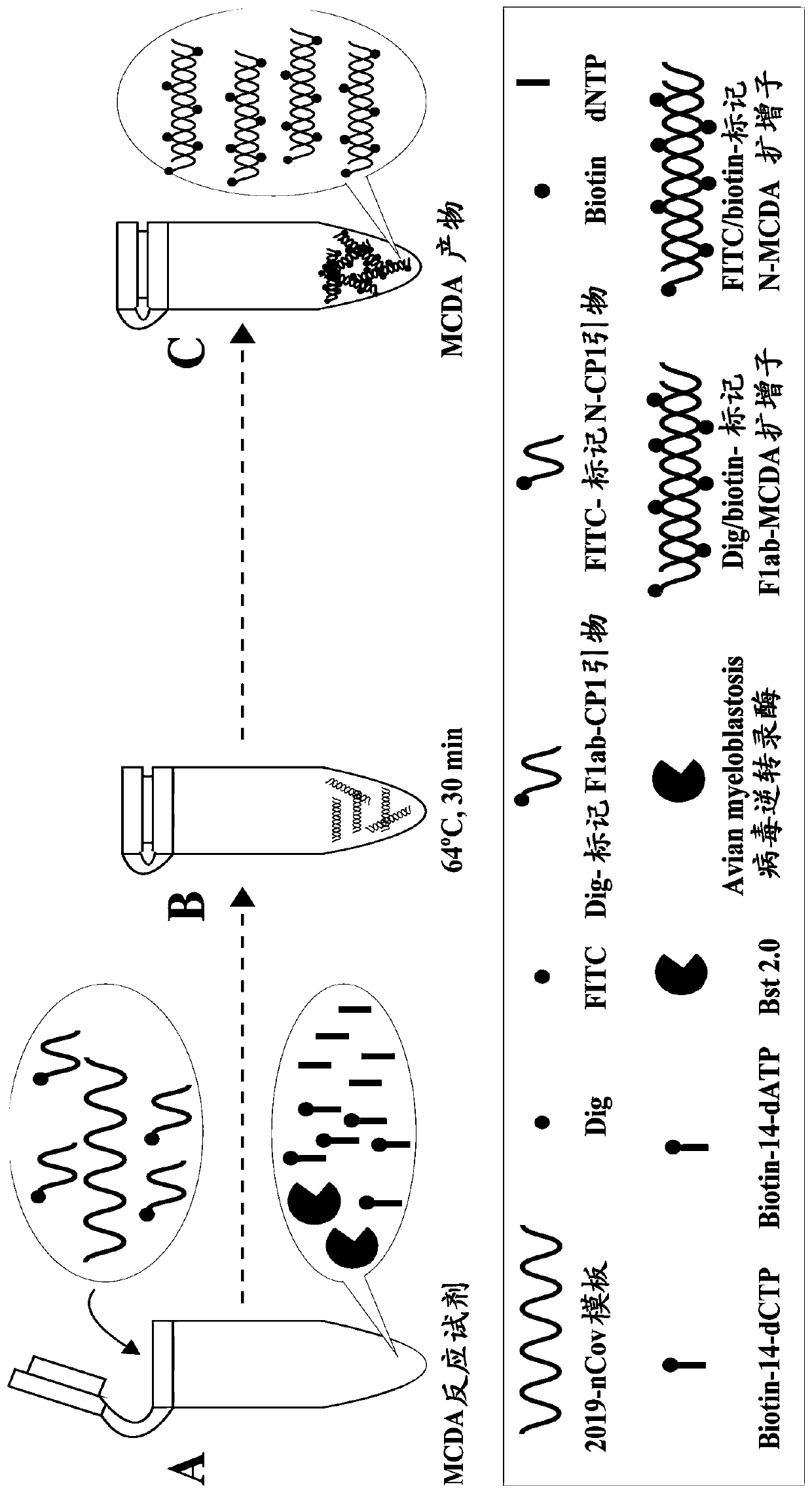
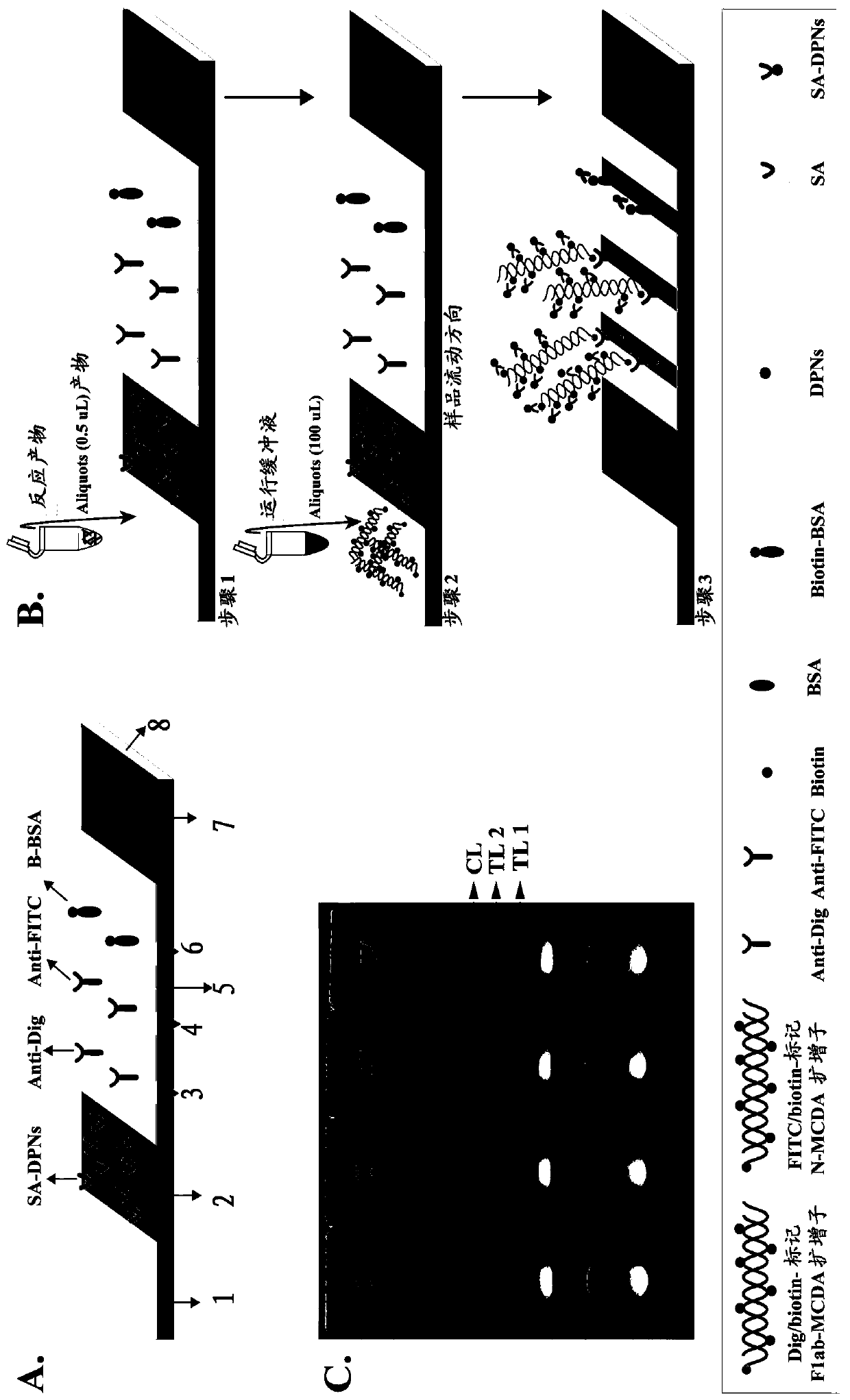
Method for detecting SARS-CoV-2 by combining reverse transcription multi-cross displacement amplification with nano-biology sensing
A cross-replacement and reverse transcription technology, applied in the field of microbiology, can solve the problems affecting the efficiency of emergency detection, false negatives, complicated operation steps, etc., and achieve the effect of excellent detection sensitivity and fast detection speed.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] The feasibility of embodiment 1.MCDA-LFB amplification
[0061] Standard MCDA reaction system: the concentration of cross primer CP1 and CP1* is 30 pmol, the concentration of cross primer CP2 is 60 pmol, the concentration of displacement primer F1 and F2 is 10 pmol, the concentration of amplification primer R1, R2, D1 and D2 is 30 pmol, The concentration of amplification primers C1 and C2 is 20pmol, 10mM Betain, 6mM MgSO 4 , 0.1 mM Biotin-14-dATP and Biotin-14-dCTP, 1.4 mM dNTP, 12.5 μL of 10×Bst DNA polymerase buffer, 10 U of strand-displacing DNA polymerase, 1 μL of template, supplemented with deionized water to 25 μl. The entire reaction was thermostated at 64°C for 35 minutes.
[0062] After MCDA amplification, the MCDA product can be detected by LFB, positive two red bands (TL and CL) appear in the detection area, negative only one red band (CL). Since the hapten labeled with the MCDA primer designed for the F1ab of SARS-CoV-2 is Dig (digoxin). Therefore, when ...
Embodiment 2
[0064] Example 2. Feasibility of Multiplex MCDA-LFB Amplification
[0065] Multiplex MCDA reaction system: the concentration of cross primer F1ab-CP1 and F1ab-CP1* is 10 pmol, the concentration of cross primer F1ab-CP2 is 30 pmol, the concentration of displacement primer F1ab-F1 and F1ab-F2 is 5 pmol, and the concentration of amplification primer F1ab- The concentration of R1, F1ab-R2, F1ab-D1 and F1ab-D2 is 15 pmol, the concentration of amplification primers F1ab-C1 and F1ab-C2 is 10 pmol, the concentration of cross primer N-CP1 and N-CP1* is 10 pmol, cross primer The concentration of N-CP2 is 30pmol, the concentration of displacement primers N-F1 and N-F2 is 5pmol, the concentration of amplification primers N-R1, N-R2, N-D1 and N-D2 is 15pmol, and the concentration of amplification primers N- The concentration of C1 and N-C2 is 10 pmol, 10 mM Betain, 6 mM MgSO4, 0.1 mM Biotin-14-dATP and Biotin-14-dCTP, 1.4 mM dNTP, 12.5 μL of 10×Bst DNA polymerase buffer, 10U of strand-dis...
Embodiment 3
[0068] Embodiment 3. determine the optimal reaction temperature of MCDA technology
[0069] Under standard reaction system conditions, add the corresponding MCDA primers designed for the SARS-CoV-2 virus F1ab plasmid and N plasmid templates, and the template concentration is 5000 copies. The reaction was carried out under constant temperature conditions (60-67°C), and the results were detected by a real-time turbidimeter, and different dynamic curves were obtained at different temperatures, see Figure 6 (F1ab sequence amplification) and Figure 7 (N gene sequence amplification). 64-65°C is recommended as the optimal reaction temperature for MCDA primers. In the follow-up verification of the present invention, 64° C. was selected as the constant temperature condition for MCDA amplification. Figure 6 and 7 Represents the temperature dynamic curve of MCDA primers designed for the detection of SARS-CoV-2 virus for F1ab and N gene sequences.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com