A method for identifying protein phosphorylation sites
A phosphorylation and protein technology, which is applied in the field of identifying protein phosphorylation sites, can solve problems such as heavy workload, and achieve the effect of speeding up the research process and saving preparation costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] This embodiment provides a method for identifying protein phosphorylation sites, which includes the following steps.
[0051] (1) Determine the phosphorylation site of the sample to be tested
[0052] Most of the possible reversible phosphorylation sites that may exist are screened through literature search or phosphoproteomics analysis.
[0053] (2) Construction of expression vectors 1 to 3
[0054] The expression vectors 1 to 3 are plasmids respectively containing amino acid sequence fragments 1 to 3, and the amino acid sequence fragments 1 to 3 are all sequence fragments containing the detection site of the protein to be detected, and the sequence lengths are the same.
[0055] Among them, the amino acid sequence fragment 1 is a fragment in which most of the possible reversible phosphorylation sites except the site to be tested are subjected to permanent non-phosphorylation site mutations on the fragment of the protein to be tested;
[0056] Amino acid sequence fra...
Embodiment 2
[0061] This embodiment provides a method for detecting phosphorylation at a single point of ATGL, and this embodiment takes the phosphorylation site of Ser47 as the research object.
[0062] (1) Screening possible phosphorylation sites of ATGL
[0063] According to literature reports, mouse ATGL has 10 phosphorylation sites modified, including Ser47, Ser87, Thr101, Thr210, Thr372, Tyr378, Ser393, Ser396, Ser406 and Ser430.
[0064] (2) Construction of expression vector
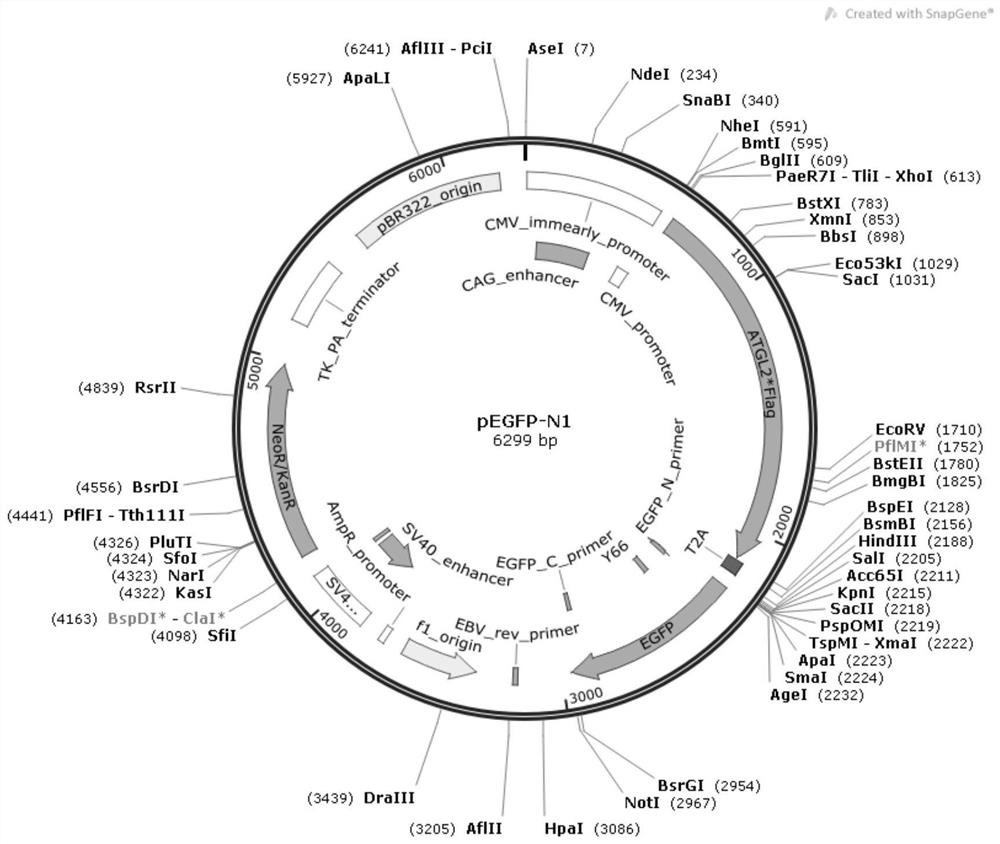
[0065] In this example, the phosphorylation site of Ser47 was used as the research object, and a plasmid with 10 permanent non-phosphorylation mutations controlled by the CMV promoter (FLAG-FLAG double tag, p9-ATGL-FLAG) was designed as a negative control , the negative control is the expression vector 2, and the ATGL vector map please refer to the attached figure 1 . The base sequence of its amino acid sequence fragment is shown in SEQ ID No.2.
[0066] The 10 permanent non-phosphorylation mutation sites ...
Embodiment 3
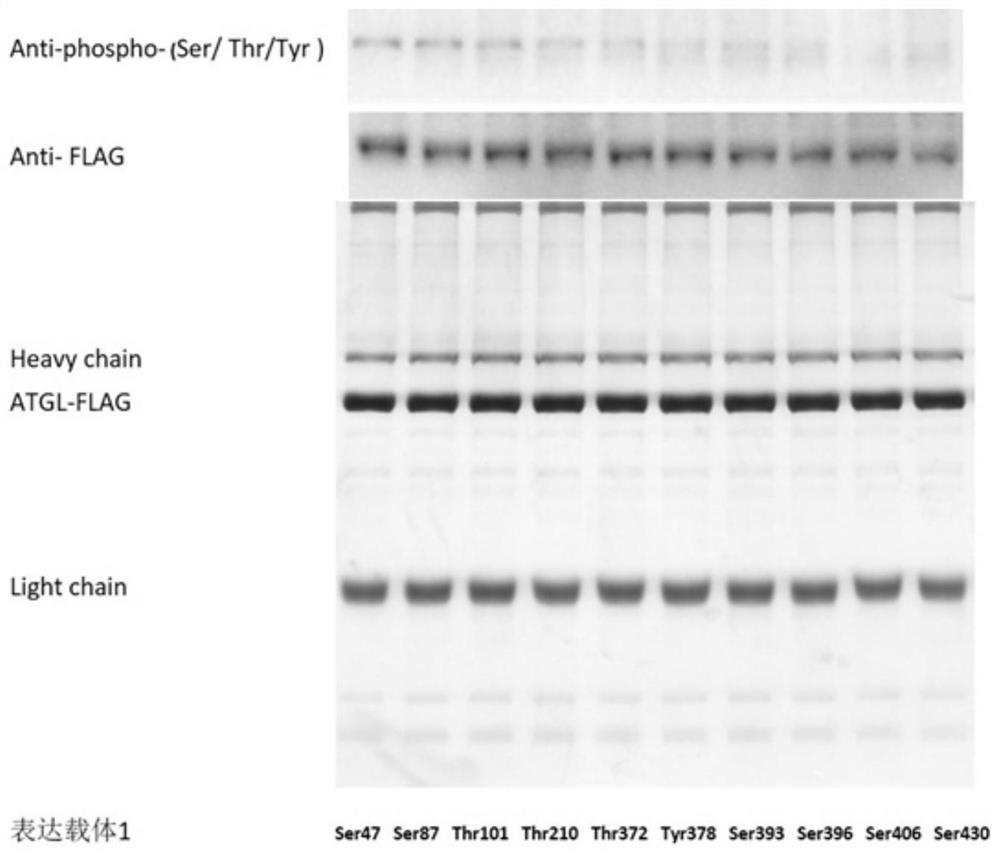
[0079] This example provides a method for detecting phosphorylation at a single point of ATGL. The specific method is the same as that in Example 2, and the phosphorylation sites of Ser47, Ser87, Thr101, Thr210, Thr372, Tyr378, Ser393, Ser396, Ser406 and Ser430 were used as the research objects.
[0080] For the above-mentioned 10 phosphorylation sites, expression vectors 1 corresponding to 10 sites to be tested were respectively prepared. Please refer to Table 1 for sequence information.
[0081] Table 1 Sequence information
[0082]
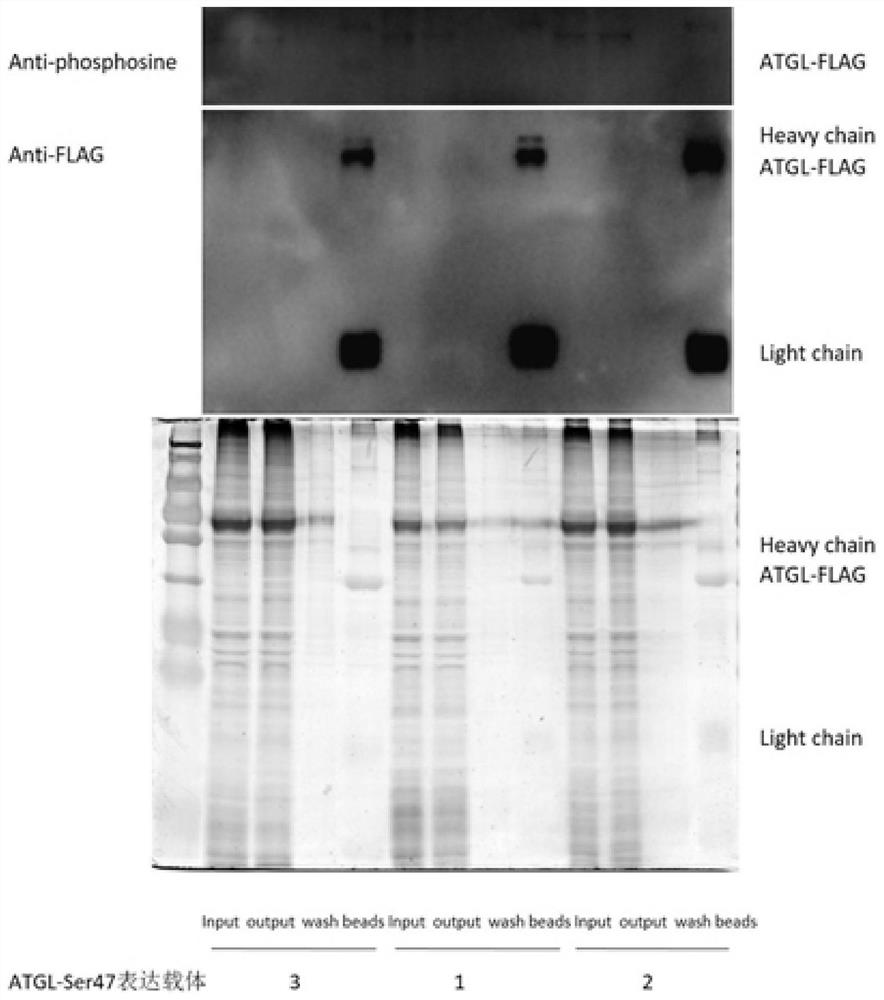
[0083] ATGL10 phosphorylation site plasmid vector 1 was transfected into HL-1 cells for 48 hours to collect protein, and FLAG magnetic beads immunoprecipitation reaction to separate ATGL protein. For results of silver staining and immunoblotting, please refer to the attached image 3 .
[0084]The results of silver staining showed that the FLAG magnetic bead binding protein was mainly ATGL protein with FLAG tag expressed by the expression ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com